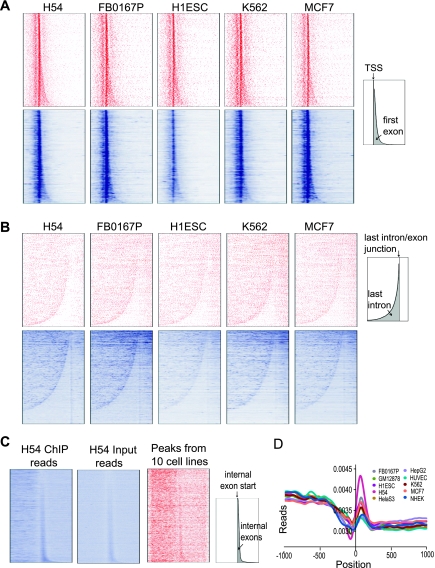
Figure 8.
RNAPII is enriched in exons. (A) Heat maps of gene-wise RNAPII occupancy. RNAPII signals in the form of input-corrected peak scores (red) or read counts (blue) were assigned to 10-bp bins across the 4-kb region shown in these plots (1 kb upstream of and 3 kb downstream from the TSS). Only genes with at least one peak or read occurrences within 4 kb of their TSS are plotted. As a result, the plotted gene sets in the top row (red) are different from that in the bottom row (blue). Five representative cell lines are shown and data for additional cell lines are in Supplemental Figure S20. The schematic at the right shows the overall gene map, with the first exon area shaded in gray. (B) RNAPII occupancy is higher in the last exon compared to the last intron. Heat maps similar to A, but genes were aligned by the start of their last exons and sorted by the length of their last introns. RNAPII signal across the 9-kb region (7 kb upstream of and 2 kb downstream from the start of the last exon) is represented similarly as in A. Most of the long genes at the bottom of the read plots (blue) do not have significant RNAPII binding and therefore are not part of the peak plots (red), a phenomenon contributing to the pattern difference between read plots and peak plots. (C) RNAPII is enriched on internal exons. Internal constitutively spliced exons were aligned by their start and sorted by exon length. RNAPII occupancy 1.5 kb upstream of and downstream from the exon start sites is plotted. In the representative H54 cell type shown, reads for both RNAPII ChIP and input showed stronger intensity within internal exons (first two panels, blue). However, the combined RNAPII peaks (input-corrected and normalized) from all 10 cell types showed modest enrichment at internal exons (third panel, red). (D) Read profiles for RNAPII ChIP-seq, after correcting for input signal using our binomial correction method. Zero indicates the start (5′ end) of constitutive internal exons.