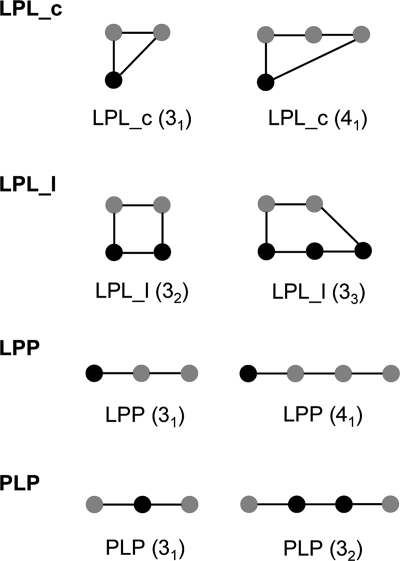
Figure 1.
Diagrams of network elements used in this study including two representative examples for each type. Black nodes denote ligand atoms, and gray nodes denote backbone amide or side chain groups of the protein reduced graph. An edge stands for a noncovalent favorable or covalent interaction. LPL_c represents ligand–protein–ligand network paths which begin and end at the same atom (ligcycle). LPL_l represents ligand–protein–ligand network paths which begin and end at different ligand atoms (ligloop). LPP represents ligand–protein–protein paths (ligpath), and PLP represents protein–ligand–protein paths. The numbers in parentheses indicate the number of nodes in the network, where connected ligand atoms are counted as one node and, in subscript, the number of connected ligand atoms in the path. Additional special network path types are derived from this collection using the following specific constraints. Privileged pairs of hydrogen bonds (HLH) are PLP elements in which the two protein–ligand contacts are hydrogen bonds with the two ligand atoms being close in space. Pure hydrogen bond networks involving neither covalent bonds nor non-hydrogen bond interactions are derived for LPL_c (LPL_chb), LPP (LPPhb), and PLP (PLPhb) network elements.