Figure 5.
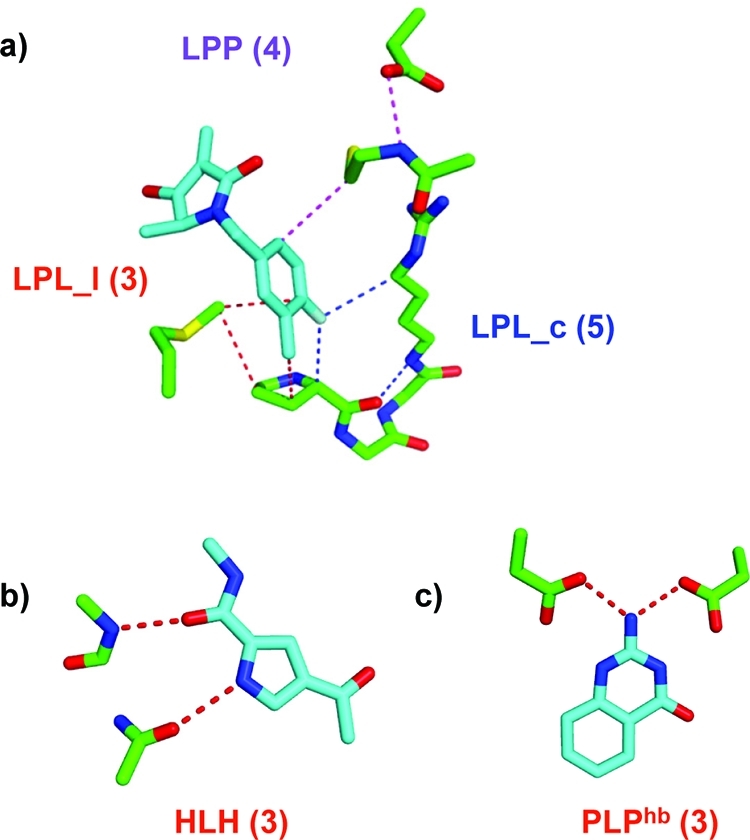
Illustration of subgraph network descriptors used in this study. The number in parentheses indicates the number of nodes in the network. Ligand atoms count as one network node, and protein residues are treated as a reduced graph with backbone amide and side chain groups counting as one node, each. (a) LPL_c is a ligand–protein–ligand network path which begins and ends at the same atom (ligcycle). LPL_l is a ligand–protein–ligand network path which begins and ends at different ligand atoms (ligloop). LPP is a ligand–protein–protein path (ligpath). (b) HLH is a privileged hydrogen bonding network element in which pairs of hydrogen bonds are adjacent to each other, and (c) PLPhb is a representative of a pure hydrogen bonding network element in which the ligand atom bridges two protein groups by hydrogen bonds. See Figure 1 and the Methods section for a complete list of network elements.
