Figure 5. Comparison between Dynamic Loop Model and self-avoiding walk.
A. The upper conformation is a Dynamic Loop Model chromatid with  , cutoff size
, cutoff size  and mean loop concentration
and mean loop concentration 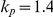 . For comparison, a conformation without loops with the same chain length
. For comparison, a conformation without loops with the same chain length  is shown below. B. For both, self-avoiding walk and Dynamic Loop Model the coarse-graining method is applied and the directional correlation is calculated. The same degree of coarse-graining is used for both models. The figure shows an exponential decay of the directional correlation function of the Dynamic Loop Model, while the the self-avoiding walk does not show this behaviour. Most importantly, the Dynamic Loop Model chromatid is much stiffer than the self-avoiding walk. This shows that the entropic repulsion of the chromatin loops that are generated by the cross-linking mechanism leads to a considerable stiffening up. Error bars represent the standard error of the sampled conformations.
is shown below. B. For both, self-avoiding walk and Dynamic Loop Model the coarse-graining method is applied and the directional correlation is calculated. The same degree of coarse-graining is used for both models. The figure shows an exponential decay of the directional correlation function of the Dynamic Loop Model, while the the self-avoiding walk does not show this behaviour. Most importantly, the Dynamic Loop Model chromatid is much stiffer than the self-avoiding walk. This shows that the entropic repulsion of the chromatin loops that are generated by the cross-linking mechanism leads to a considerable stiffening up. Error bars represent the standard error of the sampled conformations.

