Abstract
The Drosophila CNS midline glia (MG) are multifunctional cells that ensheath and provide trophic support to commissural axons, and direct embryonic development by employing a variety of signaling molecules. These glia consist of two functionally distinct populations: the anterior MG (AMG) and posterior MG (PMG). Only the AMG ensheath axon commissures, whereas the function of the non-ensheathing PMG is unknown. The Drosophila MG have proven to be an excellent system for studying glial proliferation, cell fate, apoptosis, and axon-glial interactions. However, insight into how AMG migrate and acquire their specific positions within the axon-glial scaffold has been lacking. In this paper, we use time-lapse imaging, single-cell analysis, and embryo staining to comprehensively describe the proliferation, migration, and apoptosis of the Drosophila MG. We identified 3 groups of MG that differed in the trajectories of their initial inward migration: AMG that migrate inward and to the anterior before undergoing apoptosis, AMG that migrate inward and to the posterior to ensheath commissural axons, and PMG that migrate inward and to the anterior to contact the commissural axons before undergoing apoptosis. In a second phase of their migration, the surviving AMG stereotypically migrated posteriorly to specific positions surrounding the commissures, and their final position was correlated with their location prior to migration. Most noteworthy are AMG that migrated between the commissures from a ventral to a dorsal position. Single-cell analysis indicated that individual AMG possessed wide-ranging and elaborate membrane extensions that partially ensheathed both commissures. These results provide a strong foundation for future genetic experiments to identify mutants affecting MG development, particularly in guidance cues that may direct migration. Drosophila MG are homologous in structure and function to the glial-like cells that populate the vertebrate CNS floorplate, and study of Drosophila MG will provide useful insights into floorplate development and function.
Keywords: apoptosis, central nervous system, Drosophila, ensheathment, midline glia, migration
Introduction
In both vertebrates and invertebrates, there exist diverse glial cell types that carry-out a variety of important nervous system functions. One central issue in developmental neuroscience is understanding how each glial cell type divides, migrates, undergoes apoptosis, recognizes and ensheaths axons, and acquires its distinctive functional morphology. In this manner, it can be determined which genetic pathways are shared among different glia, and which are distinct. This will be important for understanding how glia function and evolve, and, in the case of humans, provide the foundation for potential therapies for glial-based diseases.
The Drosophila midline glia (MG) are a distinct glial cell type that ensheath the two axon commissures, anterior commissure (AC) and posterior commissure (PC), that cross the midline of the CNS (Crews, 2009). There are two MG subtypes, anterior MG (AMG) and posterior MG (PMG) that have distinct gene expression patterns and interactions with the commissures (Wheeler et al., 2009; Watson et al., 2011). MG are present throughout embryonic, larval, and pupal development, but are absent from the adult CNS, suggesting that their roles are developmental (Awad and Truman, 1997). One potential function of MG is to provide trophic support to crossing axons, and the recent identification of midline-expressed neurotrophins supports this concept (Zhu et al., 2008). The MG also comprise a multifunctional embryonic signaling center controlling aspects of axon guidance, epidermal formation, muscle cell migration, and formation of the mesodermal dorsal median cells (Crews, 2003). When embryonic MG are ablated, the axon commissures become severely disorganized (Bergmann et al., 2002). This emphasizes the importance of Drosophila MG.
Insect MG are highly related to the glial-like cells that lie at the ventral midline or floorplate of the developing vertebrate nerve cord and brain. The floorplate cells also ensheath and guide commissural axons (Yoshioka and Tanaka, 1989; Campbell and Peterson, 1993; Garbe and Bashaw, 2004) and act as a critical signaling center that patterns the spinal cord by secreting Sonic hedgehog (Dessaud et al., 2008). Given the important roles of the floorplate, it is likely that floorplate-related dysfunction may lead to neurological and mental health disorders. Despite considerable knowledge regarding the signaling role of the floorplate in axon guidance and pattern formation, relatively little is known regarding the development of floorplate cells or how they ensheath commissural axons. In this regard, the Drosophila MG provide an excellent, generalizable system for the study of floorplate glia.
Previous studies on MG development have provided useful information regarding apoptosis, migration, and ensheathment (Bergmann et al., 2002; Crews, 2009). However, these studies were limited by visualizing MG in fixed embryos. In most cases, AMG could not clearly be distinguished from PMG, nor could accurate patterns of migration be discerned. In order to more fully understand MG development, we have utilized three complementary approaches for studying MG development. The first involved staining embryos with markers that provided the ability to distinguish AMG from PMG and to detect MG undergoing division and apoptosis. The second approach utilized live imaging of MG to track their movement throughout development. The third approach involved imaging individual MG in stained embryos to assess the extent of membrane elaboration and commissural axon ensheathment. Using these approaches, we have assembled a comprehensive view of MG development that will provide a strong foundation for future genetic studies.
This view incorporates a description of the migration, proliferation, and apoptosis of MG during embryonic development. We show that after their formation, both AMG and PMG migrate inward over midline neurons toward the nascent axon commissure. AMG then initiate a second posterior migration in which they move along stereotyped paths, ultimately residing in positions where they elaborate complex processes that surround and intercalate into the axon commissures. Analyses of individual AMG migration paths from time-lapse imaging indicate a correlation between AMG positions at the start of the posterior migrations with their choice of migration path and destination. Taken together, these data suggest the existence of guidance cues to direct AMG along different migration paths and the possibility that the interpretation of those cues by AMG is dictated by position. Near the end of the inward migration, a subset of AMG divides and produces, on average, 2 additional AMG per segment. Paradoxically, these and other AMG almost immediately undergo apoptosis. Apoptosis continues sporadically in both AMG and PMG until late in embryogenesis, leaving approximately 3 AMG and no PMG. The work described in this paper will be particularly useful for future studies in identifying genes that govern how glial number is regulated, how glia respond to guidance cues and migrate to specific locations around axons, and how they interact with those axons.
Materials and methods
Drosophila strains
Drosophila strains used included 380-slit-Gal4 (Wharton and Crews, 1993), sim-Gal4 UAS-tau-GFP (Wheeler et al., 2006), UAS-GFP-actin (Verkhusha et al., 1999), UAS-mCD8-GFP (Lee and Luo, 1999), UAS-moesin-GFP (Bloor and Kiehart, 2001), UAS-tau-GFP (Brand, 1995), and P[12xSu(H)bs-lacZ] (Go et al., 1998), and P{nos-phiC31\int.NLS}; P{CaryP}attP2 (Groth et al., 2004).
Generation of MG-expressed Gal4 lines
Two novel MG-expressed Gal4 transgenic lines were generated and used for either live imaging (glec-O1.5-Gal4) or single-cell analysis (argos-G1.1-Gal4). The glec-O1.5 fragment resides in the 3′ flanking sequence, and was generated by PCR amplification of genomic DNA using the forward primer 5′-TCTCCCGGAAACGAAGGAGTTCCTG-3′ and reverse primer 5′-CATAATCGTTGTCCTGTGATCCTACGTTTG-3′ (Chr 3R: 17672381–17673859; Assembly: BDGP R5\dm3). Note that glec-O1.5 spans a genomic region containing both glec MG enhancers described in Fulkerson and Estes, (2010). The argos-G1.1 fragment resides in intron 1, and was generated by PCR-amplification of genomic DNA using the forward primer 5′-GTGCACACGCACACTCAGACTCGCAC and reverse primer 5′-CTACTCTTCCAGCTTCTTCGCCAGCACAG-3′ (Chr 3L: 16466506–16467651). To clone the sli-380 fragment (Wharton et al., 1993), forward primer 5′-ATTTAAGTTGCTTGCCATGCTGGAG-3′ and reverse primer 5′-GTGAGTGACATTCCATGGGGAGC-3′ were used to PCR amplify from genomic DNA (Chr 2R: 11775792–11776179). Fragments were cloned into either pCR8 or pENTR (Invitrogen), Gateway-cloned into pBPGw-UCP, and transformed into P{nos-phiC31\int.NLS}; P{CaryP}attP2 Drosophila embryos containing the PhiC31 destination site attP2 (68A1-B2) (Groth et al., 2004) and expressing posteriorly-localized PhiC31 integrase.
Construction of the pBPGw-UCP PhiC31 transformation vector
The pBPGw-UCP plasmid is a Drosophila PhiC31-based Gal4 transformation vector in which enhancer-containing fragments are cloned adjacent to a universal core promoter (UCP) followed by Gal4. The UCP sequence was kindly provided by Jim Kadonaga. To generate the UCP insert, fragment four oligonucleotides were designed and annealed, generating the double-stranded sequence: 5′-CCGGCAGCGGTATAAAAGGGCGCGGGGTGGCTGAGAGCATCA GTTGTGAATGAATGTTCGAGCCGAGCAGACGTGCCGCTGTAC-3′; italicized nucleotides were single-stranded overhangs to match FseI and KpnI restriction site overhangs, respectively; transcription start +1 nucleotide is underlined. The Gateway-compatible PhiC31 transformation Gal4 reporter vector pBPGw (Pfeiffer et al., 2008) was cut with FseI and KpnI, and ligated to the annealed UCP fragment. The resulting pBPGw-UCP is missing 82 bases of promoter-flanking sequences compared to pBPGUw (Pfeiffer et al., 2008) due to the restriction sites used, but is otherwise functionally equivalent.
In situ hybridization, immunostaining, and microscopy
In situ hybridization and immunostaining were carried out as previously described (Kearney et al., 2004; Wheeler et al., 2006). Tyramide Signal Amplification (TSA) (Perkin Elmer) was used for indicated antibodies. Primary antibodies: mouse anti-β-galactosidase (1:500, Promega), mouse MAb BP102 (1:33, Developmental Studies Hybridoma Bank [DSHB]), rabbit anti-activated Caspase-3 (1:25 with TSA amplification, Abcam, ab13847), rabbit anti-GFP (1:500, Abcam, ab290), and rabbit anti-phospho-Histone H3 (1:250, Millipore, 06–570), guinea pig anti-Sim (1:200 with TSA) (Ward et al., 1998), and guinea pig anti-Wrapper (1:200) (Wheeler et al., 2009). The wrapper digoxygenin-labeled antisense RNA probe used for in situ hybridization was generated from cDNA clone GH03113 from the Drosophila Gene Collection (Open Biosystems). Midline cells were visualized in abdominal segments A1–A6 using Zeiss LSM Pascal and LSM-710 confocal microscopes.
Time-lapse imaging of MG
Imaging of MG migration was carried-out using the following transgenic strains: (1) glec-O1.5-Gal4 UAS-mCD8-GFP, (2), glec-O1.5-Gal4 UAS-GFP-actin, (3) glec-O1.5-Gal4 UAS-moesin-GFP, and (4) 380-slit-Gal4 UAS-mCD8-GFP. Embryos were collected for 3 hours at room temperature and aged for 14 hours at 18°C. They were then dechorionated for 2 minutes in 50% bleach, mounted on glass coverslips, and covered with halocarbon oil 700 (Sigma, H8898). Coverslips were placed oil-side down on PetriPERM 50mm hydrophobic cell culture dishes (Sarstedt, 96077305) that had an oxygen-permeable membrane. GFP fluorescent images were captured using a Zeiss LSM-710 confocal microscope with a 40X oil-immersion objective. Embryos were visualized beginning at mid to late stage 12 until stages 15–16 (approximately 6 hours) or from stages 15 to 17 (approximately 4 hours). Imaging was generally terminated when the embryo rotated, thus moving the midline out of the plane of focus. Z-series stacks were captured every 2 minutes. For movies, individual focal planes were chosen for each time point using Zeiss Zen 2009 software, then aligned, annotated, and converted into movies using Adobe Photoshop.
Tracking and mapping of MG position
To track the position of midline cells from time-lapse imaging experiments, the center of the nucleus of individual cells was marked relative to the anterior face of either the fused single commissure (prior to separation) or the AC (after separation) using Zen 2009. Measurements were made for each 50 minute interval. The intervals were shortened if apoptosis or rapid migration occurred. Using Adobe Illustrator, single images from each measured time point were aligned such that the anterior and ventral faces of the AC were in register between time points. Magnifications were normalized across experiments, and colored circles were placed over the position marks. The AC from each experiment was aligned to the AC in schematics of idealized segments to generate maps of MG position over time.
Results
Inward migration of MG during stages 11–12
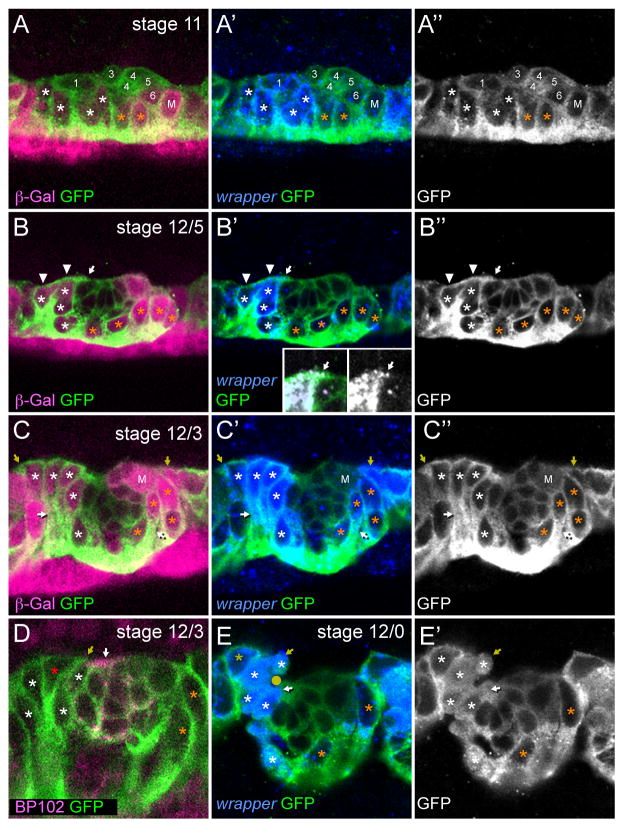
Our analyses during stages 11 and 12 relied on two markers that distinguish: (1) AMG from PMG and (2) MG from midline neurons. There are approximately 18 embryonic midline neurons in each segment that arise from 6 midline neuronal precursors (MPs): MP1, MP3, MP4, MP5, MP6 and the median neuroblast (MNB) (Wheeler et al., 2008). The first marker, wrapper RNA, is found in both AMG and PMG from embryonic stages 10–17, and transiently in MP1 prior to its division at stage 11 (Fig. 1A) (Wheeler et al., 2008; Watson et al., 2011). wrapper expression is present at high levels in AMG and low levels in PMG. The second MG marker is a transgene, 12xSu(H)bs-lacZ, which is a reporter of Notch signaling (Wheeler et al., 2008). During stage 10, Delta-Notch signaling activates MG gene expression, including 12xSu(H)bs-lacZ. Reporter expression is present at stages 11–12 in AMG, PMG, as well as MP5, MP6, and the MNB, which are Notch-dependent midline neural precursors. The reporter is found at higher levels in PMG compared to AMG (Fig. 1A–C). These markers were analyzed in embryos also harboring single-minded (sim)-Gal4 UAS-tau-GFP, which is present in the cytoplasm of all midline cells. In these embryos, the cytoplasm appears as a brightly stained ring around an unstained nucleus, and allows visualization of MG cell shape, process extension, and nuclear morphology.
Fig. 1. Inward migration of MG.
Fluorescence confocal images of single segments in sagittal view from sim-Gal4 UAS-tau-GFP embryos. Anterior is left, dorsal (internal) is up and ventral (external) is down. White asterisks indicate AMG and orange asterisks indicate PMG. Not all MG present in each segment are shown, because they were in different focal planes. RNA (italicized) and proteins (non-italicized) stained and their corresponding colors are indicated in the lower left corner. Embryonic stage is indicated in the upper right corner. (A–C″) MG were identified by visualizing expression of 12xSu(H)bs-lacZ (anti-β-galactosidase), wrapper RNA, and cell shape (anti-GFP). (A–A″) During stage 11, MG are elongated along the dorsal-ventral axis and are located ventral to the midline neurons and MPs. At this stage, MP1 (1) has not yet divided, MP3 (3) is in the process of dividing, MP4 has already divided into 2 VUM4 neurons (4), and MP5 (5), MP6 (6), and MNB (M) have not yet divided. In contrast to MG, MP1 is elongated along the anterior-posterior axis. 12xSu(H)bs-lacZ is expressed at low levels in AMG and MP5 and at high levels in PMG, MP6, and MNB (M). At this stage, wrapper is expressed at high levels in AMG and MP1 and at low levels in PMG. (B–B″) By stage 12/5, AMG have begun moving internally around the anterior of the midline neurons. The most internal AMG (arrowheads) orient towards the posterior. In some segments, wrapper+ AMG cytoplasm from one or more AMG (arrows) extends to the posterior over the top of the midline neurons. The insets in B′ show a magnified view of the region containing the wrapper+ AMG cytoplasmic extension. For clarity, wrapper is shown in white with and without GFP. (C–C″) By stage 12/3, most AMG have migrated internally and PMG have begun to migrate inward, posterior towards the MNB (M). Both AMG and PMG have long trailing processes that extend to the surface of the embryo (white arrows). The most anterior AMG often orient their dorsal-most membranes towards the anterior (yellow arrows). (D) At stage 12/3, there is a single commissure (white arrow) that stains with MAb BP102 and resides above the midline neurons. An AMG oriented towards the posterior (red asterisk) extends cytoplasm (yellow arrow) towards the commissure. (E,E′) By stage 12/0, most AMG have migrated inward and no longer have trailing processes. AMG begin to extend wrapper+ cytoplasm both above (yellow arrow) and below (white arrow) the single commissure. The commissure can be recognized as the unstained space surrounded by AMG cytoplasm (yellow circle in E). A subset of AMG show nuclear and cytoplasmic localization of tau-GFP (white asterisks) indicating that they are in the process of dividing.
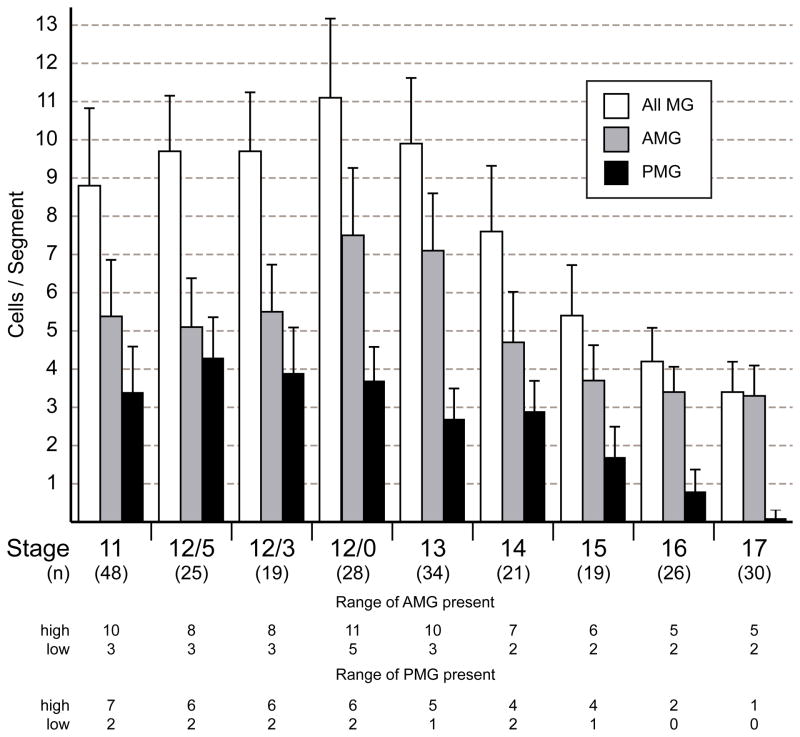
Confocal imaging of wild-type embryos with these MG-staining reagents allowed analysis of the number, position, and movement of MG from stage 11 to the end of stage 12. At stage 11, MG, distinguished by large nuclei, were elongated along the dorsal-ventral axis (Fig. 1A). The MP1 also transiently appeared large and elongated prior to its division, but it was extended along the anterior-posterior axis (Fig. 1A). During stage 11, MG were positioned both below and anterior to the cluster of developing MPs and midline neurons (Fig. 1A). The total number of MG (wrapper+ 12xSu(H)bs-lacZ+) could be counted, and partitioned into AMG (high wrapper, low 12xSu(H)bs-lacZ) and PMG (low wrapper, high 12xSu(H)bs-lacZ). Often MG in the middle of the segment showed low levels of both wrapper and 12xSu(H)bs-lacZ. These cells were considered late-differentiating AMG based on their low levels of 12xSu(H)bs-lacZ and presence of runt, an AMG marker (not shown) (Watson et al., 2011). At stage 11, there were 8.8 ± 2.0 MG in each segment (n=48 segments analyzed) (Fig. 2), although this number was highly variable ranging from 6 to 15 cells/segment. Of these MG, 5.4 ± 1.4 (n=48) were AMG ranging from 3 to 10 cells and 3.4 ± 1.2 (n=48) were PMG ranging from 2 to 7 cells (Fig. 2). The variability in AMG and PMG numbers did not show obvious correlations either within segments or between neighboring segments (Table 1). Segments with large numbers of PMG did not have corresponding smaller numbers of AMG nor did adjacent posterior segments have reduced numbers of AMG.
Fig. 2. Variation in the number of MG during embryogenesis.
Comparison of the number of AMG, PMG, and total MG from stages 11 to 17 from the analysis of stained confocal images. The number of segments examined (n) is noted below each stage. Data are means ± standard error of the means. At stages 11 to 12/0, all MG were counted using the expression of 12xSu(H)bs-lacZ and wrapper. AMG were high wrapper and low 12xSu(H)bs-lacZ and PMG were low wrapper and high 12xSu(H)bs-lacZ. From stages 13 to 17, MG were identified by position and expression of wrapper, (high in AMG and low in PMG). The numbers of AMG and PMG are relatively constant from stages 11 to 12/3. From stages 12/0 to 17, average AMG number spikes before declining to approximately 3 AMG/segment while the number of PMG declines to near 0 by stage 17. At the bottom, the range of values for the number of AMG or PMG at each stage is shown as the highest (high) and lowest (low) number counted.
Table 1.
Relationship between the number of AMG and PMG during stage 11
| # of AMG | n | AMG/segment | P-value | |
|---|---|---|---|---|
| Total AMG | 257 | 48 | 5.4 ± 1.4 | -- |
| AMG from same segment | ||||
| 2 PMG | 57 | 11 | 5.2 ± 1.1 | 0.71 |
| 3 PMG | 91 | 18 | 5.1 ± 1.3 | 0.45 |
| 4 PMG | 72 | 12 | 6.0 ± 1.9 | 0.29 |
| > 4 PMG | 37 | 7 | 5.3 ± 1.4 | 0.97 |
| AMG from next posterior segment | ||||
| 2 PMG | 45 | 8 | 5.6 ± 1.2 | 0.62 |
| 3 PMG | 56 | 12 | 4.7 ± 1.0 | 0.13 |
| 4 PMG | 40 | 8 | 5.0 ± 1.7 | 0.53 |
| > 4 PMG | 25 | 4 | 6.3 ± 1.7 | 0.25 |
The top row (Total AMG) indicates the total number of AMG and mean AMG/segment counted from 48 segments (n). AMG from same segment – the number of AMG were counted in 48 segments that contained varying numbers of PMG. For each group differing in PMG number, the mean, standard deviation, and P-value were calculated for the number of AMG present in the same segments. AMG from next posterior segment – 32 segments had an adjacent posterior segment. For each group differing in PMG number, the mean, standard deviation, and P-value were calculated for the number of AMG present in the next posterior segments. The Student’s t-test with a 2-tailed distribution was used to compare the mean AMG/segment between each experimental class and the total mean AMG. None of the experimental values differed significantly from the total AMG mean.
Embryonic stage 12 is a major period of MG migration, and this was analyzed in detail in confocal images of stained embryos. CNS morphogenesis is rapid during stage 12, and is commonly divided into three substages (12/5, 12/3, 12/0) based on the progression of germband retraction (Klämbt et al., 1991). Embryos proceed from stage 11 to 12/5, 12/3, 12/0, and then 13 (the first number before the “/” is the stage and second number refers to the number of segments that still reside on the dorsal side of the embryo). During stage 12/5, the morphology of AMG began to change (Fig. 1B). AMG with nuclei close to the dorsal surface reoriented towards the posterior, often extending membrane along the top of the midline neurons towards the position of the nascent single commissure (Fig. 1B,D). Between stages 12/5 and 12/3, most AMG and PMG began migrating internally (Fig. 1B,C). AMG migrated inward anterior to the midline neurons while PMG migrated inward posterior to the midline neurons. One feature of the inward migration was that both AMG and PMG trailed long processes that remained associated with the underlying ectodermal cells (Fig. 1C). Not all AMG oriented towards the posterior. The most anterior AMG oriented towards the anterior (Fig. 1C) indicating differences in migratory behavior between AMG. From stages 12/3 to 12/0, AMG continued to migrate internally and lost their trailing processes (Fig. 1E). A subset of AMG began migrating to the posterior both above and below the single commissure (Fig. 1E). This posterior movement ultimately results in the ensheathment of the axon commissures by AMG (see below). At the same time that posterior migration began, the localization of tau-GFP in a subset of AMG changed from strictly cytoplasmic to both cytoplasmic and nuclear (Fig. 1E′). Previously, we associated this change in tau-GFP localization with cells undergoing mitosis (Wheeler et al., 2008).
AMG divide and undergo apoptosis
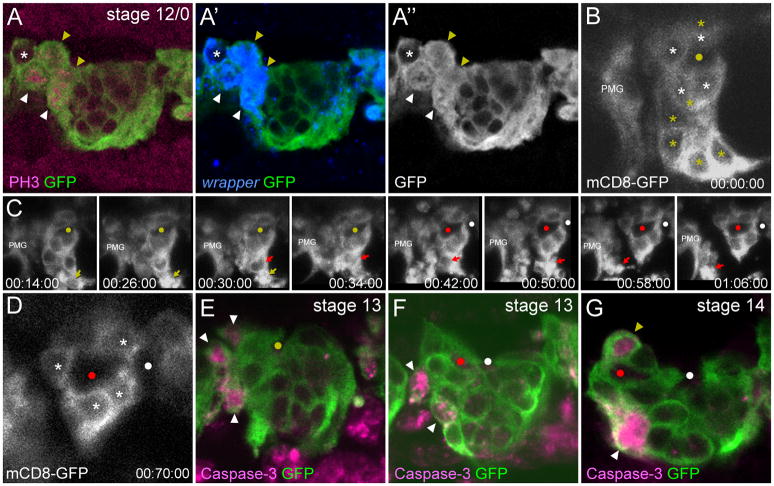
The change in tau-GFP localization indicated that AMG were dividing at stage 12/0. This was confirmed by anti-phospho-histone-H3 (PH3) staining. In these experiments, 90% of segments showed at least 1 division with an average of 1.9 PH3+ AMG per segment (n=60) (Fig. 3A). The addition of approximately 2 AMG per segment due to division is consistent with the observed increase in the number of AMG from 5.5 ± 1.2 at stage 12/3 to 7.5 ± 1.7 at stage 12/0 (Fig. 2). AMG divisions occurred at stage 12/0 as no PH3+ AMG were observed at stage 12/3 (n=54 segments) and only 1 PH3+ AMG was observed at stage 13 (n=43 segments). In contrast, no PH3 staining was observed in PMG.
Fig. 3. AMG divisions and apoptosis.
(A–A″) Sagittal view of a single segment from a stage 12/0 sim-Gal4 UAS-tau-GFP embryo containing 5 AMG (asterisk and arrowheads). Anterior is to the left. Four wrapper+ AMG show nuclear and cytoplasmic tau-GFP (arrowheads). Of these, 2 are PH3+ (white arrowheads) and 2 are PH3− (yellow arrowheads). The PH3− cells are likely just beginning division prior to large scale phosphorylation of Histone H3. (B–D) Individual frames taken from a time-lapse imaging experiment of a glec-O1.5-Gal4 UAS-mCD8-GFP embryo at stage 13 in sagittal view focused around the AC. PMG from this segment are not shown, but PMG from the next anterior segment are present (PMG). Yellow circles indicate the single commissure prior to separation, red and white circles indicate the AC and PC respectively after separation, and elapsed time is shown (hh:mm:ss). (B) Initially, there are 10 AMG in this segment (asterisks). The 4 AMG that are in contact with the AC (white asterisks) will survive. The remaining AMG (yellow asterisks) will migrate inward and undergo apoptosis leaving a space between AMG and PMG from the next anterior segment. (C) Over the course of 52 minutes, 6 AMG fragment, and the debris is rapidly cleared. The yellow and red arrows indicate the progression of 2 AMG undergoing apoptosis. (D) After the wave of cell death, only 4 AMG remain (white asterisks). (E–G) Single segments from (E,F) stage 13 and (G) stage 14 sim-Gal4 UAS-tau-GFP embryos in sagittal view stained with anti-active Caspase-3 and anti-GFP. Yellow circle in (E) marks the position of the single commissure prior to separation, and red and white circles in (F) and (G) indicate the AC and PC, respectively. (E,F) During stage 13, AMG located at the anterior of the segment, and not in contact with the commissures, are active Caspase-3+ (white arrowheads). The position along the dorsal-ventral axis of active Caspase-3+ AMG varies. (G) During the period of sporadic AMG apoptosis (stage 14–16), both AMG that are at a distance from the AC (white arrowhead) and AMG that appear to contact the commissure (yellow arrowhead) are active Caspase-3+.
Between stages 13 (7.1 ± 1.5 AMG/segment) and 14 (4.7 ± 1.3 AMG/segment), the number of AMG was reduced by an average of 2.4 cells per segment (Fig. 2). To visualize the dynamics of AMG cell death, the membrane marker UAS-mCD8-GFP was expressed under the control of an MG-specific enhancer derived from the gliolectin (glec) locus and imaged in live embryos. glec-O1.5-Gal4 UAS-mCD8-GFP fluorescence was first observed during stage 12/0 in AMG and at a lower level in PMG. Fluorescence did not appear sufficiently early to visualize AMG divisions, but did allow imaging of AMG cell death. Nine segments were analyzed in time-lapse experiments, and in 5 segments, the reduction in AMG was dramatic and occurred as a wave of cell death during stage 13. In one example, there were 11 AMG in a segment; 5 of these fragmented and the debris was quickly cleared, leaving 6 surviving AMG (Fig. 3B-D; Supplemental Movie 1). In earlier studies, MG fragments were observed in embryonic macrophages indicating that they phagocytose and clear MG corpses (Sonnenfeld and Jacobs, 1995). Based on our time-lapse imaging, segments with higher numbers of AMG at stage 12/0 underwent a wave of cell death during stage 13. In segments in which a wave of apoptosis occurred, AMG number was reduced from an average of 9.8 AMG/segment to 5.2 AMG/segment (n=5). Segments that did not undergo a wave of apoptosis had fewer AMG at stage 12/0, only 7.2 AMG/segment (n=4). Previous studies showed that the reduction in MG is due to apoptotic cell death (Sonnenfeld and Jacobs, 1995; Dong and Jacobs, 1997; Zhou et al., 1997; Bergmann et al., 2002). Consistent with this, a subset of AMG during stages 12/0 to 13 possessed activated Caspase-3 indicating activation of the apoptotic pathway (Fig. 3E–G). Similar to apoptotic AMG positions observed in time-lapse imaging, activated Caspse-3+ AMG were found at the anterior margin of the segment, distant from the AC.
AMG migrate posteriorly and extend projections into the commissures
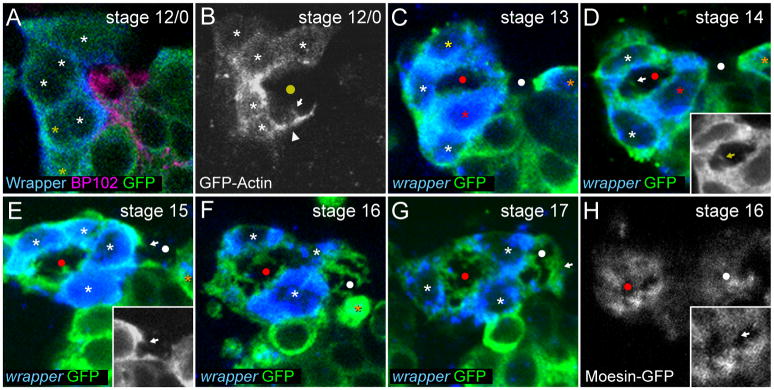
At stage 12/0, AMG were arrayed along the anterior side of the segment and began to interact with the nascent axon commissure (Fig. 4A). While some AMG were undergoing apoptosis, others began to surround the commissure. These AMG sent GFP-actin-containing processes above and below the commissure (Fig. 4B). The processes were followed shortly after by the posterior migration of AMG nuclei (Fig. 4C). At this point, the single commissure separated into the AC and PC (Klämbt et al., 1991). AMG cytoplasm moved into the space between the commissures and contacted AMG cytoplasm approaching from the opposite side (Fig. 4C). Shortly after, an AMG nucleus was observed in the space between the AC and PC (Fig. 4D). By stage 15, 2–3 AMG nuclei were present above and 1–2 below the AC (Fig. 4E), with cytoplasm surrounding the AC but not the PC. The number and position of AMG nuclei did not change significantly between stages 15 and 17 (Figs. 2, 4F,G), but there was continued development of the AMG projections.
Fig. 4. AMG surround the axon commissures and extend membrane projections.
Sagittal views of single segments from (A,C–G) stained sim-Gal4 UAS-tau-GFP embryos, and time-lapse imaging of (B) glec-O1.5-Gal4 UAS-GFP-actin and (H) glec-O1.5-Gal4 UAS-moesin-GFP embryos focused on the AC and the surrounding AMG. A yellow circle marks the position of the single commissure while red and white circles mark the AC and PC respectively. (A) At stage 12/0, Wrapper+ AMG (asterisks) are arrayed along the anterior of the segment. Some AMG (white asterisks) are in contact with the AC (BP102+) while others are not (yellow asterisks). (B) In live embryos at stage 12/0, AMG send actin-GFP-containing processes under (white arrowhead) and into (white arrow) the commissure. (C) During stage 13, AMG migrate posteriorly above (yellow asterisk) and below (red asterisk) the AC. AMG cytoplasm has extended around the AC from both sides, ensheathing the AC. PMG (orange asterisk) contact the posterior of the PC. (D) During stage 14, an AMG migrates between the commissures (red asterisk). At the same time, tau-GFP-containing AMG projections (white arrow) become visible in the AC. Inset shows tau-GFP only and is focused on the AC. PMG remain in contact with the posterior of the PC. (E) During stage 15, AMG nuclei surround the AC and AMG begin to send projections into the PC (white arrow). Inset shows tau-GFP only and is focused on the PC. One PMG remains in contact with the posterior of the PC but does not ensheath. (F) In a stage 16 embryo, AMG send elaborate projections into both the AC and PC. In this segment the last PMG has undergone apoptosis leaving an apoptotic body (orange asterisk). (G) During stage 17, AMG projections into the PC become more elaborate and AMG cytoplasm surrounds the PC (white arrow). (H) At stage 16 in live embryos, moesin-GFP is localized to the regions of AMG that make contact with the commissures including AMG projections. Inset is focused on the AC, white arrowhead indicates moesin-GFP+ AMG projections.
AMG projections into the AC were first observed by tau-GFP visualization during stage 14 as small protrusions from AMG cell bodies (Fig. 4D). In addition to microtubules, AMG projections also contained GFP-actin (Fig. 4B) and moesin-GFP, an actin-binding ERM protein (Fehon et al., 2010). Moesin-GFP was localized only to AMG projections, not throughout the cell (Fig. 4H). This indicates that AMG projections are likely specialized domains distinct from other AMG compartments. Interestingly, it has been shown that the Canoe and Shotgun (E-cadherin) proteins are also highly localized in and around these projections and could play a role in their formation or maintenance (Slovakova and Carmena, 2011). The outgrowth of AMG projections may begin as early as stage 12/0 since small GFP-actin containing protrusions were observed at this stage (Fig. 4B). By stage 15, tau-GFP+ AMG projections were readily observed in the AC and just beginning to enter the PC (Fig. 4E). From stages 15 to 17, the AMG projections increased in size and complexity (Fig. 4F,G). Eventually, at stage 17, the posterior face of the PC was completely covered by AMG membrane (Fig. 4G), and both AC and PC possessed extensive MG membrane projections.
AMG migrate along stereotypical paths to surround the anterior commissure
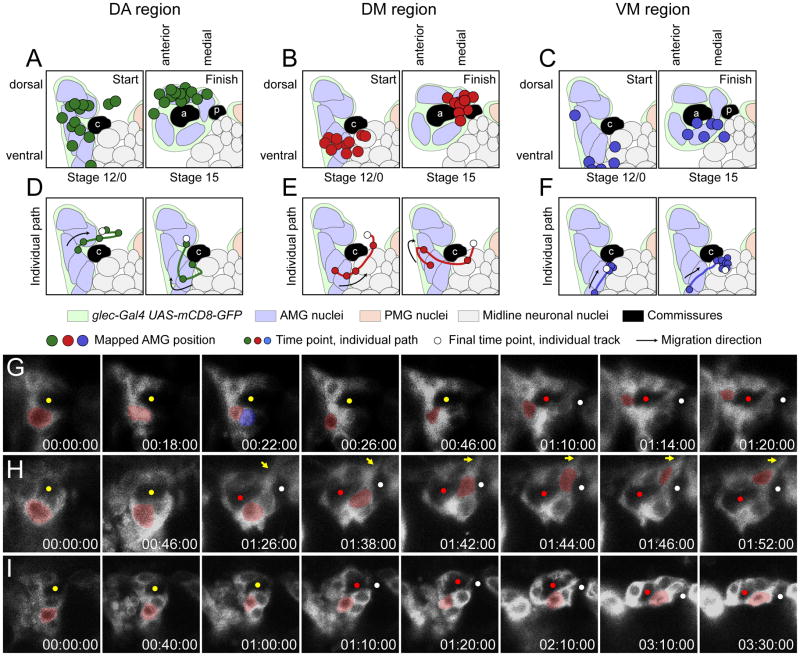
To better understand the dynamics of AMG migration, time-lapse imaging was used to track the movement of AMG over time. We compared MG visualization by driving expression of UAS-mCD8-GFP with either 380-slit-Gal4 or glec-O1.5-Gal4, two MG-expressed Gal4 lines we derived from the slit and gliolectin (glec) genes. glec-O1.5-Gal4 showed higher levels of mCD8-GFP and was used for these studies. The membrane marker mCD8-GFP allowed the visualization of cell membranes and nuclei (dark areas within GFP+ membranes). Since glec-O1.5-Gal4 was expressed in all AMG, we could not discern the membranes of individual AMG and instead examined the movement of individual nuclei. Post-imaging analyses of AMG migration and apoptosis were done digitally by marking nuclei at stage 12/0 and tracing their movements until the end of imaging, usually stage 15 (Fig. 5). A total of 66 AMG from 10 segments were tracked. In each segment, approximately half of the AMG underwent apoptosis (see below) leaving 3–4 surviving AMG per segment, a total of 31 cells. To investigate the correlation between initial and final positions of surviving AMG, the starting and finishing positions of individual AMG migration paths were mapped onto a schematic of an idealized midline segment. Additional imaging from stages 15–17 showed that surviving AMG do not migrate further (data not shown) such that AMG position at stage 15 is an accurate indicator of their final position. From these analyses, trends in AMG position and migration were identified. The AMG located most dorsally (internal) at the beginning of the analysis tended to end their migration dorsally, above the AC (Fig. 5A). We refer to this as the dorsal-anterior (DA) region. AMG that began migration from an intermediate position within the segment also migrated to a dorsal position, but in between the commissures (Fig. 5B). This is the dorsal-medial (DM) region. AMG beginning migration from the most ventral (external) positions tended to cease their migration below and in between the commissures (Fig. 5C) – this is the ventral-medial (VM) region. Because of variability in the exact position of AMG at stage 15, the 3 regions were defined based on their relative position. For example, a dorsally-located AMG that resides between the AC and PC may be assigned to the DA region if there is another dorsally-located AMG that is more posterior (this one would be assigned to DM). Thus, there is overlap in position between the posterior-most AMG assigned to the DA region and the anterior-most AMG assigned to the DM region (Fig. 5A,B).
Fig. 5. AMG migrate along stereotypical paths.
(A–C) Schematics of idealized stage 12/0 and stage 15 segments focused on the commissures and the surrounding AMG. The starting and finishing positions of 31 AMG from time-lapse imaging experiments are mapped onto these idealized segments (see Materials and methods). At stage 12/0 there is a single commissure labeled c, and at stage 15 the AC and PC are labeled a and p, respectively. Other symbols are defined in the key. (A) Most AMG that completed their migration above the AC (DA region) started their migration in the most dorsal group of midline cells at stage 12/0 (14 AMG analyzed). (B) AMG that ceased migration above and between the commissures (DM region) began their migration in the intermediate region at stage 12/0 (11 AMG analyzed). One circle is completely obscured by other overlapping circles. (C) Most AMG that migrated to the region below and between the commissures (VM region) started their migration in the most ventral portion of the segment (6 AMG analyzed). (D–F) Two examples of individual AMG migration paths for each position. Paths are drawn on a schematic of stage 12/0, but represent migration during stages 12/0 to 15. (G–I) Time-lapse experiments showing the paths of individual AMG that migrated to the (G) DA, (H) DM, and (I) VM regions. The AMG of interest is pseudocolored in red and elapsed time is shown (hh:mm:ss). Yellow circles mark the single commissures; red and white circles mark the AC and PC. (G) The AMG begins migration below the commissure, divides (red and blue pseudocolor), and migrates internally to the DA region, adjacent to the AC. (H) An AMG pauses for 1 hour and 26 minutes before rapidly (6 min) migrating internally between the commissures to the DM region. Yellow arrow indicates a long process that extends above the midline, presumably emanating from the migrating AMG. (I) This AMG migrates from an external position to contact the AC and stays in the VM region for the remainder of the experiment.
How do AMG migrate from the anterior of the segment at stage 12/0 to their final positions? Analyses of individual migration paths showed that most surviving AMG followed one of three common paths. For AMG that migrated to the DA region (n=14), they started in the most dorsal position at stage 12/0, migrated more internally in some cases, and then to the posterior (Fig. 5A,D,G; Supplemental Movie 2). Migration was not always direct, as AMG sometimes migrated back to a more anterior position. AMG that migrated to the VM region (n=6) started in the most ventral position and migrated posteriorly and inward towards the commissure (Fig. 5C,F,I; Supplemental Movie 3). This movement often occurred in the first 60 minutes of migration (5/6 cells). By contrast, DA region AMG often migrated for more than 2 hours before becoming stationary (8/14 cells). Once VM AMG contacted the commissure in the VM region, they ceased to migrate further. In contrast to the relatively straightforward migrations of other AMG, DM region AMG began migration from an intermediate position and migrated posteriorly beneath the AC (n=11). Often they would pause in this position before rapidly moving up between the separated commissures to the dorsal side (Fig. 5B,E,H; Supplemental Movie 4). Occasionally, AMG attempted to migrate between the commissures but were blocked by AMG that were already present in that position. Thus, it is likely that AMG migrating towards the DM region are the AMG that are often observed between the commissures in stained samples (Fig. 4D). Due to the fact that all AMG and PMG were labeled by glec-O1.5-Gal4 UAS-mCD8-GFP, it was difficult to discern the membrane shape of individual AMG. However, due to the relative lack of other MG along the path of AMG migrating to the DM region, a long process was sometimes observed (3/11 cells) (Fig. 5H; Supplemental Movie 4) that likely emanates from these AMG cells. In all cases, the processes went between the commissures and extended out towards a point above the midline (Fig. 5E,H).
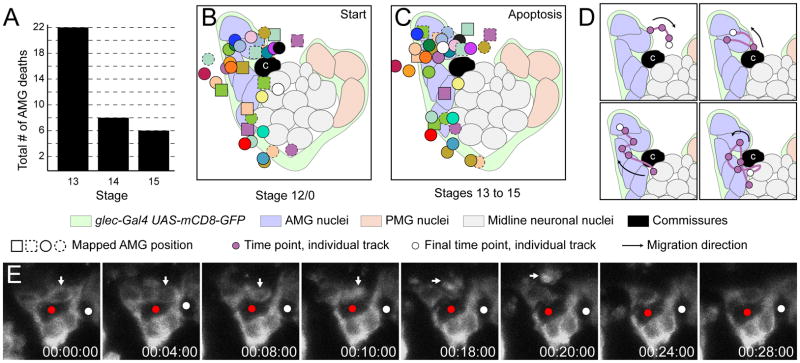
From stages 12/0 to 15, the number of AMG was reduced to an average of 3.7 AMG/segment (Fig. 2). No additional waves of apoptosis were observed, but individual AMG sporadically underwent apoptosis with most cells dying during stage 13 (Fig. 6A,E; Supplemental Movie 5). Unlike the earlier waves of apoptosis, many of these AMG were in close proximity to the anterior commissure (Fig. 3G,6E). For these dying cells, starting positions were similar to the positions in which they underwent apoptosis (Fig. 6B,C). Analysis of individual migration paths indicated that most AMG migrated a short distance prior to apoptosis. Others migrated long distances sometimes returning close to their starting points (Fig. 6D).
Fig. 6. AMG undergo sporadic apoptosis during stages 13–15.
(A) Chart showing the number of AMG that underwent apoptosis at stages 13–15 from a total of 35 AMG assayed in time-lapse imaging experiments. Stages were determined by elapsed time from stage 12/0; stage 13 – 0–60 min, stage 14 – 61–120 min, stage 15 – 121–220 min. (B,C) Schematics of idealized stage 12/0 segments with the single commissure labeled “c”. The position of AMG are mapped (B) at the start of imaging and (C) at the time of apoptosis as assayed by membrane fragmentation. All observed apoptoses occurred during stages 13 to 15. Apoptosis position is mapped on to stage 12/0 to facilitate comparison with (B). The symbol for each AMG is designated by a combination of shape, color, and border (plain or dashed). Other symbols are defined in the key. (B) AMG that ultimately did not survive are present along the anterior of the segment during stage 12/0. (C) AMG undergo apoptosis at many positions, but do not migrate far from their position as the start of imaging. (D) Four examples of individual AMG migration paths that ended with the AMG fragmentation and subsequent clearance of the cellular debris. Paths are shown with schematics of stage 12/0, but represent paths from stages 12/0 to 15. (E) Time-lapse experiment showing the apoptosis and clearance of a single AMG during stage 13 (white arrow). Another AMG immediately takes its place. Red and white circles mark the AC and PC. Elapsed time is shown (hh:mm:ss).
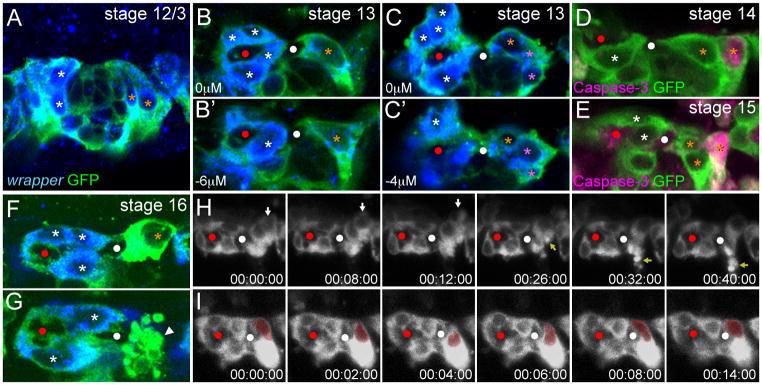
PMG migrate inward, contact the posterior commissure, and undergo apoptosis
The inward migration of PMG was analyzed using stained embryos due to the low level of glec-O1.5-Gal4 UAS-mCD8-GFP initially present in PMG. The paths of PMG migration are distinct from those of AMG. Instead of orienting to the posterior as do most AMG, PMG orient in an anterior direction towards the commissure (Fig. 7A). After commissure separation, PMG elongated along the anterior-posterior axis and contacted the posterior face of the PC (Fig. 7B). Often two PMG were observed with this position and elongated morphology (Fig. 7B′). Additional PMG had more rounded nuclear morphologies and formed a cluster of cells posterior to the elongated PMG (Fig. 7C). PMG underwent apoptosis sporadically and were completely absent by the middle of stage 17 (Fig. 2). Like AMG, apoptosis first occurred in PMG at a distance from the commissure (Fig. 7D,E,H; Supplementary Movie 6) indicating that interaction with the commissure may transiently protect PMG from apoptosis. During stage 16, the morphology of PMG adjacent to the PC changed; the nucleus became more rounded and the cytoplasm more intensely tau-GFP+ or mCD8-GFP+ (Fig. 7F,I). Interestingly, we observed two stage 16 PMG nuclei that repeatedly extended towards the commissure and then retracted (Fig. 7I; Supplemental Movie 7). This may indicate that PMG position is a balance between attractive and repulsive cues from neighboring cells. Based on staining, the last PMG in each segment underwent apoptosis between the end of stage 16 and the middle of stage 17 (Fig. 7G). After the last PMG was removed, the PC was surrounded by AMG membrane (Fig. 4G).
Fig. 7. PMG migrate, contact the posterior commissure, and undergo apoptosis.
(A–C,F,G) Single segments from sim-Gal4 UAS-tau-GFP embryos in sagittal view stained for wrapper RNA and anti-GFP. AMG are labeled with white asterisks and PMG with orange asterisks. Red and white circles mark the AC and PC. (A) During stage 12/3, PMG elongate and migrate internally. Not all AMG or PMG are present in the focal plane shown. (B,B′) Single segment of a stage 13 embryo showing 2 PMG in different focal planes. These PMG are elongated along the anterior-posterior axis and make contact with the posterior of the PC. (C) Two focal planes showing a group of PMG (pink asterisks) with rounded nuclear morphologies that reside in the posterior of the segment, and don’t contact the PC. (D,E) PMG that do not contact the commissure sporadically become active Caspase-3+. (F,G) During stage 16, (F) PMG cytoplasm becomes intensely GFP+ and (G) some PMG undergo apoptosis leading to presence of apoptotic bodies (white arrowhead). (H,I) Time-lapse experiments showing: (H) the apoptosis of a single PMG (white arrow) during stage 15 leading to the presence of apoptotic bodies (yellow arrow), and (I) the repeated internal and external movement of a PMG nucleus (red pseudocolor). Elapsed time is shown (hh:mm:ss).
Individual AMG reveal extensive membrane elaboration
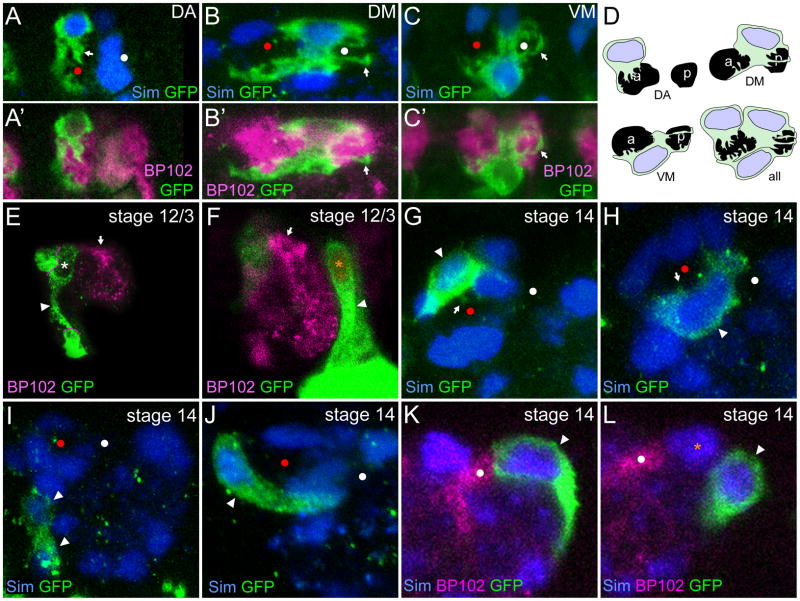
Since AMG follow particular tracks and occupy stereotyped positions with respect to the commissures, we investigated how AMG that reside in the three different regions interact with the commissures. This required analyzing individual AMG. This was accomplished using a transgenic strain containing argos-G1.1-Gal4 combined with UAS-tau-GFP that expresses GFP sporadically in 1–2 AMG or PMG in each segment. At stages 16 and 17, single AMG in each region were imaged in stained embryos. AMG from the DA region covered the anterior and dorsal faces of the AC (Fig. 8A) and had projections that extended almost exclusively into the AC (6/7 cells). AMG in the DM region (n=9) covered portions of both the AC and PC and had elaborate projections into both (Fig. 8B). Likewise, AMG from the VM region (n=16) covered portions of both the AC and PC and had projections extending into both commissures (Fig. 8C). Projections from AMG in the DM and VM regions often spanned the entire length of a commissure, while those from AMG in the DA region were more restricted. In no case (n=25) did a single AMG completely surround the entire AC or PC. Thus, the AC and PC are surrounded by a combination of AMG membranes from multiple AMG that extend around from both the dorsal and ventral sides (Fig. 8D). The region between the commissures was covered by AMG from both the DM region (9/9 cells) and the VM region (11/16 cells). Thus, in most segments, some regions of the commissural surface were covered by multiple AMG while other regions were only covered by a single AMG.
Fig. 8. Individual MG have distinct positions and morphologies.
Individual AMG or PMG from argos-G1.1-Gal4 UAS-tau-GFP embryos in sagittal view. Red and white circles show the AC and PC. Anterior is to the left and dorsal is up. (A–C′) The elaboration of projections and cell shape (anti-GFP) for individual AMG during stage 16 is shown for AMG in the: (A,A′) DA region, (B,B′) DM region, and (C, C′) VM region. Anti-Sim (blue) staining reveals MG nuclei in (A–C), and MG and a subset of midline neurons in (G–L). MAb BP102 axonal staining shows the location of the commissures. (A,A′) Sim+ AMG nucleus resides above the AC in the DA region, and GFP staining reveals that its cytoplasm covers the anterior face of the AC and portions of the dorsal and ventral faces. Projections (white arrow) are present only in the AC. (B,B′) An AMG nucleus resides above and between the AC and PC in the DM region. Cytoplasm covers portions of the dorsal face of the AC and PC and extends between the commissures covering portions of the ventral face of both the AC and PC. Projections enter both the AC and PC with some extending the length of the PC (white arrow). (C,C′) The nucleus of an AMG resides in the VM region, under and between the AC and PC. The cytoplasm covers portions of the ventral faces of the AC and PC, extends up in between the AC and PC, and covers portions of the dorsal faces of the AC and PC. Projections are found in both commissures with some extending the length of the PC (white arrow). (D) Schematic interpretation of individual AMG morphology at stages 16 and 17 in the DA, DM, and VM regions based on images from 25 AMG. Together, AMG cover the entire surface of both commissures. (E,F) At stage 12/3, individual (E) AMG and (F) PMG have long trailing processes (white arrowheads) as they migrate internally. The single commissure is shown by anti-BP102 staining (white arrow). Dotted lines in E demarcate the boundaries with GFP-stained non-midline cells. (G–L) At stage 14, there are distinct morphologies of (G–J) AMG and (K,L) PMG. (G,H) AMG that are in contact with the AC (white arrowhead) stain strongly for GFP, have an elongated cuboidal shape, and show small projections into the commissures (white arrows). (I) AMG at a distance from the commissure stain weakly with GFP and are more rounded (white arrowheads). (J) An AMG (white arrowhead) has an extended comet-like morphology that does not contact the epidermis. (K) At stage 14, a PMG (white arrowhead) contacting the PC has an elongated shape and retains a trailing process. (L) In contrast, a PMG that does not touch the PC (white arrowhead) is rounded and resides posterior to a PMG (orange asterisk) that does contact the PC. No PMG were identified at any stage that ensheathed or sent projections into the commissures.
At earlier stages, the morphology of MG was distinct from later stages. At stage 12, most inward migrating AMG oriented posteriorly towards the commissure and trailed long processes that ended in the epidermis (Fig. 8E). Similarly, inward migrating PMG trailed long processes to the epidermis but oriented towards the commissure in an anterior direction (Fig. 8F). At stage 14, AMG were observed around the AC having a roughly cuboidal shape and the beginnings of projections (Fig. 8G,H) while others were more rounded, smaller, weakly GFP-stained, and showed no projections (Fig. 8I). These morphological differences likely reflect AMG that will survive (cuboidal shape) versus those that will undergo apoptosis (rounded shape). Individual AMG with long comet-like shapes were observed during stage 14 (Fig. 8J). This morphology is similar to the AMG that move between the commissures en route to the DM region. Alternatively, all migrating AMG may extend processes followed by nuclear migration.
By examining individual PMG at stage 14, differences in the morphology of the PMG that contact the PC and those that do not could be distinguished. PMG that contacted the PC were elongated along the anterior-posterior axis and often retained their trailing processes (Fig. 8K). In contrast, PMG that did not contact the PC were more rounded and did not have trailing processes (Fig. 8L). Note that even individual PMG that contact the PC do not extend processes or ensheath the commissure, even transiently. This is consistent with the characterization of AMG as ensheathing glia and PMG as distinct, non-ensheathing glia (Watson et al., 2011).
Discussion
New imaging methods and novel insights into MG development
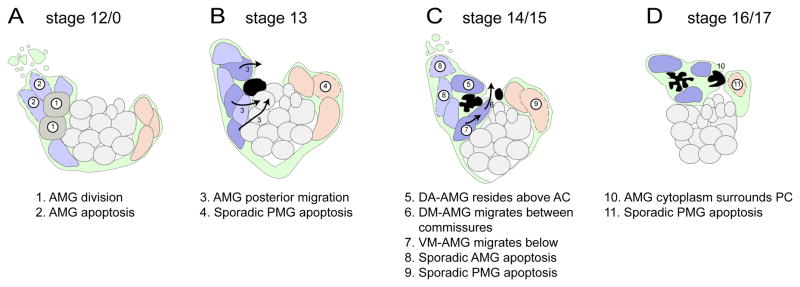
While cellular analysis of Drosophila MG development has been studied for >20 years, novel insights were gained in this study through the use of multiple advanced imaging approaches. The most important was the use of time-lapse imaging. This allowed tracking of each MG cell between stages 12 and 17, and revealed their stereotyped migration. Development at earlier times (stages 11 to 12) was assessed by analysis of sim-Gal4 UAS-tau-GFP embryos stained for MG markers. Finally, the visualization of individual MG using argos-G1.1-Gal4 UAS-tau-GFP embryos allowed the extensive cytoplasm/membrane extensions of each AMG to be determined. In addition, the analyses of MG in sagittal view allow for a simplified and more accurate reconstruction of MG migration compared to traditional ventral views. Combined, these approaches resulted in a comprehensive cellular view of MG development (summarized in Fig. 9) that will be the basis for future genetic analyses.
Fig. 9. Summary of MG migration, apoptosis, and axon ensheathment.
(A–D) The schematic depicts 4 periods of embryonic development showing the locations of AMG (nuclei colored blue and gray), midline neurons (light gray), PMG (tan), and commissural axons (black). MG membrane/cytoplasm is depicted in green. Dotted lines indicate nuclei of MG undergoing apoptosis. The dark blue AMG survive and constitute the mature stage 16/17 MG, whereas the lighter blue AMG undergo apoptosis. The AMG in gray at stage 12/0 will undergo division. Arrows depict migration pathways. Numbers indicate prominent developmental events described below the illustrations.
AMG commitment and cell division
Using multiple cell markers and confocal analysis, we were able to accurately count the number of AMG and PMG throughout development. Overall, our numbers agree well with our previous work (Wheeler et al., 2006) and that of Dong and Jacobs (1997). There are ~8 sim+ mesectodermal cells/segment at embryonic stage 7. During stage 8, these cells synchronously divide during the mitotic cycle 14 division to give rise to ~16 cells/segment (Foe, 1989). We hypothesize that these 16 cells are initially committed to become neural precursors, and subsequent Notch signaling acts via lateral inhibition to partition these cells into a population of neural precursors and MG (Wheeler et al., 2008). This process results in 8.8 MG/segment (5.4 AMG and 3.4 PMG) at stage 11. The number of AMG and PMG remain constant through stages 12/5 and 12/3 but between stages 12/3 and 12/0 there is an increase of 2.1 AMG/segment to 7.5, suggesting two divisions on average from existing AMG. Consistent with this view is the presence of PH3+ tau-dense AMG at stage 12/0. However, it is worth noting that AMG number (and presumably divisions) can vary significantly between segments. Thus, mechanistically, AMG can be generated in two ways. First, lateral inhibition mediated by Notch signaling leads to the formation of 5.4 AMG/segment during stages 10 and 11; a recruitment step that likely occurs without cell division. Second, divisions of existing AMG occur at stage 12/0 to generate a total of 7.5 AMG/segment (Fig. 9A).
Both ecdysone and vein (vn) signaling may play roles in AMG cell division. Steroids are known to influence glial proliferation and apoptosis in vertebrates (e.g. Zhang et al., 2002). Genetic and in vitro studies also indicate that high levels of ecdysteroids can inhibit proliferation and promote apoptosis of Drosophila embryonic and postembryonic MG (Awad and Truman, 1997). These results suggest that AMG have a tendency to divide, but high levels of ecdysone inhibit division. In the embryo, there is a prominent pulse of ecdysone that peaks at 8 hours of development (stage 12) and gradually decreases until hatching (Maróy et al., 1988). Thus, the presence of high levels of ecdysone may prevent AMG proliferation during late embryogenesis, but may not increase quickly enough to prevent AMG from undergoing division during stage 12. Consistent with this view, mutants of genes involved in ecdysone production (e.g. disembodied, shadow, spook) result in additional embryonic MG (Giesen et al., 2003). While the presence of ecdysone may inhibit MG proliferation, vn signaling may be required for MG divisions. The vn gene encodes a secreted ligand that binds to Egfr (Schnepp et al., 1996) and is a homolog of vertebrate neuregulin, which promotes glial division (Canoll et al., 1996). vn was previously hypothesized to control AMG division at stage 12, since vn mutant embryos showed a reduction of 2.6 AMG/segment (Lanoue et al., 2000). This is similar to the average increase in AMG due to cell division (2.1 AMG/segment). If vn is required for AMG division throughout development, then division should be absent (regardless of ecdysone levels) when vn expression is absent. This is indeed the case as vn is expressed in the midline and surrounding cells from stages 10 to 12, but is restricted to only a small subset of lateral CNS neuroblasts and neurons after AMG divisions (Lanoue et al., 2000). Taken together, this suggests that AMG proliferation may depend on a balance between positive (vn) and negative (ecdysone) signaling.
AMG apoptosis
AMG number peaks at stage 12/0 (7.5/segment) and declines by apoptosis to 3.3 AMG/segment by stage 17. This reduction in number has been well-studied and a model proposed as follows (Bergmann et al., 2002). AMG begin to express a pro-apoptotic gene, head involution defective (hid) beginning at stage 13. The axon commissures secrete Spitz (Spi), which is an EGF-like protein. If sufficient Spi protein binds to Egfr located on the surface of AMG, MAP kinase is activated, which then phosphorylates and inactivates Hid, thereby blocking apoptosis. Thus, it is predicted that AMG in close proximity to the axon commissures will receive sufficient Spi to survive, and those not in close contact will undergo apoptosis.
Time-lapse imaging provided additional insight into AMG apoptosis. This process often occurred in a two-step manner. In segments with a large number of AMG (~10) there was a rapid wave of apoptosis that yielded ~5 AMG (Fig. 9A). Further reduction of AMG to ~3 cells occurred incrementally (Fig. 9C). In segments with a smaller number of AMG (~7), all AMG underwent apoptosis incrementally with no detectable apoptotic wave. Generally, the AMG that underwent apoptosis in initial waves were those at the anterior margin of the segment, furthest from the axon commissures. This is consistent with those cells receiving less axonal Spi. While all AMG cell death is likely due to inadequate Spi signal, time-lapse imaging experiments revealed instances in which AMG in close proximity to the commissures died, were cleared, and another AMG quickly assumed its position. Thus, even cells in apparent contact with axon commissures may not receive sufficient Spi for survival, suggesting additional complexity in the regulation of AMG apoptosis.
Previously we argued that there might be a positive feedback loop in which Spi signaling from axons results in increased adhesion between AMG and axons (Crews, 2009). MG and axons adhere to each other via the heterophilic adhesion membrane proteins, Wrapper (on MG) and Neurexin IV (Nrx-IV) (on axons) (Stork et al., 2009; Wheeler et al., 2009). Ensheathment and membrane projection into axons by AMG requires wrapper function. Expression of a wrapper enhancer is upregulated when Spi is provided ectopically (Estes et al., 2008). This suggests that Spi could promote stronger adhesion and more elaborate contacts between AMG and axons via increased Wrapper levels. Increased adhesion and surface area contact could expose AMG to higher concentrations of Spi making survival even more likely. In this way, AMG would compete for axonal interactions, with advantage going to those with more elaborate contacts.
PMG apoptosis
There are ~4 PMG/segment at stage 12, and we found no evidence from PH3 staining or time-lapse imaging that PMG divide after their initial recruitment and specification by Notch and Hedgehog signaling. PMG number gradually declines to zero from stage 12/5 to stage 17 (Fig. 9B–D). The PMG furthest from the PC die first. This suggests that the PMG adjacent to the PC may transiently receive a survival signal, although they too die between stages 16 and 17. It is generally thought that: (1) PMG do not share a large surface area with commissural axons (perhaps, in part due to low levels of Wrapper; (Watson et al., 2011)), (2) their survival cannot be rescued by high levels of Spi (Bergmann et al., 2002), and (3) their apoptosis is promoted by a different combination of proapoptotic genes than AMG (Zhou et al., 1997). However, a detailed mechanistic analysis of PMG cell death has not been carried-out, but is now feasible.
Patterns of MG migration
By combining staining of early embryos with time-lapse imaging of older embryos, we were able to define in great detail the pattern of MG migration from stages 10 to 17. The AMG at stage 11 initially are present along the anterior/posterior axis in the anterior half of the segment and lie beneath (more external to) midline neurons and their precursors. The AMG nuclei begin to delaminate and migrate inward (Fig. 9A). Some AMG migrate internally and orient in a posterior direction towards the developing axon commissure (Fig. 9B). One possibility is that AMG initiate migration in response to and towards developing commissural axons or the MP1 neurons that lie just beneath the developing commissure. If axons provide guidance cues, migration may be due to an axon-derived secreted morphogen. If so, it is unlikely to be Spi, since in embryos in which both Egfr signaling is abolished and apoptosis inhibited, MG migration appears relatively normal (Bergmann et al., 2002). Another possibility is the PDGF- and VEGF-related factors (Pvfs) that may be secreted by axons or midline neurons and activates the PDGF and VEGF-related receptor (Pvr) (Learte et al., 2008). Alternatively, MG could survey axonal membrane proteins at a distance using gliopodia, which are filopodia-like membrane extensions that act like growth cone filopodia (Vasenkova et al., 2006). PMG also migrate inwards towards the developing axons and may be responding to similar guidance cues as AMG.
Not all AMG orient towards the developing axons. The anterior-most AMG elongate inwards, but tend to orient towards the adjacent anterior segment before undergoing apoptosis (Fig. 9A). This suggests that they are either blocked from responding to the normal AMG guidance cues, possibly by other AMG, or are intrinsically different from the AMG that migrate in the posterior direction. Regarding intrinsic differences, most AMG-expressed genes we have analyzed at stages 11 and 12 are expressed in all AMG, and not in AMG subsets (Wheeler et al., 2006), indicating that obvious subsets of AMG are not apparent. One exception are the medial late-differentiating AMG present at stage 11 that are runt−(Watson et al., 2011). However, after stage 11, these AMG are indistinguishable from other AMG, and it is uncertain whether their migratory paths are distinct. Consequently, it remains possible that different AMG subpopulations exist with intrinsic migratory differences, but there is currently little evidence to support this view.
Time-lapse imaging has provided new and definitive information regarding how individual MG acquire their positions surrounding the axon scaffold. One of the key observations was that the final migratory paths and positions of AMG correlated with their location at stage 12. AMG do not move randomly, they instead migrate in specific ways around the axon commissures to take their final positions. This suggests that AMG at different locations respond to specific, yet different, cues that guide them to their final positions. In addition, some mechanism must ensure that AMG cease their migration. The signals for both guidance and the termination of migrations might include a combination of AMG-AMG interactions, as well as interactions (either diffusible or contact-mediated) between AMG and different neurons, axons, or other substrata. As demonstrated by single-cell analysis, each AMG that ensheaths the commissures has extensive membrane projections that, in principle, can interact with other AMG and potential substrates. In both Nrx-IV and wrapper mutants, MG fail to adhere to the commissures and nearby neuronal cell bodies (Noordermeer et al., 1998; Stork et al., 2009; Wheeler et al., 2009). However, AMG still migrate inward, migrate posteriorly to surround the AC, and maintain adhesion among themselves. This suggests that different adhesive interactions are required for different aspects of AMG migration.
The inward migration of MG suggests that there are 3 distinct groups of MG that may undergo collective guidance (AMG that migrate inward and anterior, AMG that migrate inward and posterior, and PMG that migrate inward and anterior) (Fig. 9A). While each MG within these groups likely adheres to other group members, it is unknown whether the groups collectively signal to each other and whether they respond collectively or individually. However, after their initial inward migration, surviving AMG and PMG migrate to characteristic positions (Fig. 9B–D), indicating a transition to more individual behavior. Nevertheless, the surviving AMG remain in close contact with each other. Most importantly, this study provides the background for genetic screens that will identity and functionally analyze genes involved in AMG migration and commissure ensheathment.
Comparisons of descriptions of MG migration
Through the use of time-lapse and stained embryo imaging, the data presented in this paper provide a definitive descriptive view of MG migration. These results are consistent with and greatly extend earlier studies on MG development and migration. However, previous workers proposed detailed models regarding MG specification and migration, and, at this point, it is worth comparing our current view of MG development to earlier versions.
One influential and detailed model (Klämbt et al., 1991) has a number of features: (1) there are 3 mature pair of MG that ensheath the commissures, and are named MGA (anterior), MGM (middle), and MGP (posterior) based on their positions along the anterior/posterior axis, (2) these MG are derived from 3 discrete MG precursors that form prior to stage 8 and divide once yielding the 6 mature MG, (3) MGP migrate in an anterior direction into the next segment where they take up residence as the most posterior MG, (4) the MGM migrate above the AC and send projections that follow along the axons of the VUM neurons (medial tract), extending between the developing AC and PC, thus separating the commissures, and (5) the MGA migrate only a short distance, residing beneath the AC and PC. Important refinements of this model proposed later by other workers (e.g. Bergmann et al., 2002) established that generally ~10 MG were generated, most underwent apoptosis, and that a group of posterior-residing MG underwent apoptosis and did not contribute to the ensheathment of the commissures (Dong and Jacobs, 1997; Wheeler et al., 2006). These posterior glia are equivalent to PMG in our model.
With respect to the position, identity, and migration of the MGA, MGM, and MGP precursors and their progeny, Klämbt et al., (1991) employed lineage tracing of enhancer trap reporters reportedly from stage 16 to stage 8. However, since expression of the MG enhancer trap lines used does not begin expression until stage 12, tracing the origins of these cells back to stage 8 could not be directly determined, and was, presumably, conjectured. Subsequent work has demonstrated that MG do not form until at least stage 10 via Notch signaling (Wheeler et al., 2008), and that Hedgehog signaling at the same stage specifies AMG and PMG cell fates (Watson et al., 2011). Thus, the identities and positions of specific subtypes of MG cannot be identified before stage 10. Additionally, in Klämbt et al., (1991) no molecular markers were described that allowed MGA and MGM to be distinguished, so the proposed migration pathways that each of these MG took to reach their final destinations could not have been derived from stained samples.
More complicated are MGP and their relationship to PMG. In some respects, these cells appear equivalent. However, all PMG undergo apoptosis, and thus are not equivalent to MGP, which were initially proposed to survive and ensheath the PC. We also note that the PMG delaminate, migrate, and die within the same segment (Fig. 9A–D). We see no evidence that they migrate from a more posterior segment (as proposed for MGP). It is possible that, in the absence of specific AMG and PMG markers, inward migrating AMG that orient toward the anterior were mistaken for migrating MGP.
Finally, regarding the issue of commissure separation, the previous model proposed that MGM tracked on the medial axon tract between the commissures, resulting in their separation. The possibility that MG may track along the medial tract to separate the commissures is an interesting idea, but has not been critically tested. We find, from single-cell analysis, that both DM region and VM region AMG extend membrane/cytoplasm between the AC and PC, indicating that more than one AMG are usually positioned between the commissures. Together, these two cells ensure that the two commissures remain distinct and separate.
Supplementary Material
A wave of apoptosis reduces AMG number
Time lapse imaging of a single segment from a glec-O1.5-Gal4 UAS-mCD8-GFP embryo in sagittal view. Anterior is to the left and dorsal is up. Movie corresponds to 1 hour and 14 minutes of development beginning during stage 13. In this segment, there are 11 AMG, not all AMG are in the focal plane. The AMG that are contacting the AC (ac) survive while the remainder fragment into apoptotic bodies and are cleared. Four individual AMG are indicated with colored arrows.
AMG migration to the DA region
Time lapse imaging of a single segment of a glec-O1.5-Gal4 UAS-mCD8-GFP embryo in sagittal view. Anterior is to the left and dorsal is up. Movie corresponds to 1 hour and 18 minutes of development beginning during stage 13 when only a single commissure (com) is present. Yellow arrow indicates the nucleus of an AMG that divides and then migrates dorsally to a position adjacent to the AC.
AMG migration to the VM region
Time lapse imaging of a single segment from a glec-O1.5-Gal4 UAS-mCD8-GFP embryo in sagittal view. Anterior is to the left and dorsal is up. Movie corresponds to 1 hour and 36 minutes of development beginning during stage 12/0. The single commissure is labeled “com”. Yellow arrow indicates the nucleus of an AMG that migrates dorsally to contact the AC.
AMG migration to the DM region
Time lapse imaging of a single segment from a glec-O1.5-Gal4 UAS-mCD8-GFP embryo in sagittal view. Anterior is to the left and dorsal is up. Movie corresponds to 32 minutes of development beginning during stage 14. Yellow arrow indicates the nucleus of an AMG that has paused below the AC (ac) before migrating between the AC and PC (pc). The AMG has a long process that extends posteriorly above the PC.
An individual AMG undergoes fragmentation and is cleared
Time lapse imaging of a single segment from a glec-O1.5-Gal4 UAS-mCD8-GFP embryo in sagittal view. Anterior is to the left and dorsal is up. Movie corresponds to 40 minutes of development beginning during stage 13. The AC and PC are labeled ac and pc. A single AMG (yellow arrow) that is in contact with the AC undergoes fragmentation and the debris is cleared.
PMG undergo fragmentation and are cleared
Time lapse imaging of a single segment from a glec-O1.5-Gal4 UAS-mCD8-GFP embryo in sagittal view. Anterior is to the left and dorsal is up. Movie corresponds to 1 hour and 8 minutes of development beginning during stage 14. The AC and PC are labeled “ac” and “pc”. A single PMG (yellow arrow) fragments and the debris is subsequently cleared. After fragmentation, cellular debris moves both dorsally (yellow arrow) and to the posterior (white arrow).
PMG nucleus extends towards and retracts from the PC
Time lapse imaging of a single segment from a glec-O1.5-Gal4 UAS-mCD8-GFP embryo in sagittal view. Anterior is to the left and dorsal is up. Movie corresponds to 52 minutes of development beginning during stage 16. The AC and PC are labeled “ac” and “pc”. PMG nucleus (yellow arrow) repeatedly extends towards and retracts from the PC.
Highlights.
Multiple imaging approaches comprehensively describe Drosophila CNS midline glial development.
Different groups of midline glia collectively migrate in different directions.
Midline glia stereotypically migrate to specific positions surrounding the axon commissures.
Suggests the existence of specific cues that guide the migration of each midline glial cell.
Individual midline glia extend elaborate membrane processes.
Acknowledgments
We thank Stephanie Stagg and Tony Perdue for assistance with time-lapse microscopy experiments, the Developmental Studies Hybridoma Bank (University of Iowa) for monoclonal antibodies, the Bloomington Drosophila Stock Center, Nasser Rusan and Mark Peifer for Drosophila strains, and Jim Kadonaga, Barret Pfeiffer, and Gerry Rubin for advice and plasmids. This work was supported by NIH grants RO1 NS64264 (NINDS) and R37 RD25251 (NICHD) to S.T.C. The UNC Developmental Biology NIH training grant, T32 HD046369 and NRSA F32 HD061175 (NICHD) provided support to JCP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Awad TA, Truman JW. Postembryonic development of the midline glia in the CNS of Drosophila: proliferation, programmed cell death, and endocrine regulation. Dev Biol. 1997;187:283–97. doi: 10.1006/dbio.1997.8587. [DOI] [PubMed] [Google Scholar]
- Bergmann A, Tugentman M, Shilo BZ, Steller H. Regulation of cell number by MAPK-dependent control of apoptosis: a mechanism for trophic survival signaling. Dev Cell. 2002;2:159–170. doi: 10.1016/s1534-5807(02)00116-8. [DOI] [PubMed] [Google Scholar]
- Bloor JW, Kiehart DP. zipper Nonmuscle myosin-II functions downstream of PS2 integrin in Drosophila myogenesis and is necessary for myofibril formation. Dev Biol. 2001;239:215–228. doi: 10.1006/dbio.2001.0452. [DOI] [PubMed] [Google Scholar]
- Brand A. GFP in Drosophila. Trends Genet. 1995;11:324–325. doi: 10.1016/s0168-9525(00)89091-5. [DOI] [PubMed] [Google Scholar]
- Campbell RM, Peterson AC. Expression of a lacZ transgene reveals floor plate cell morphology and macromolecular transfer to commissural axons. Development. 1993;119:1217–1228. doi: 10.1242/dev.119.4.1217. [DOI] [PubMed] [Google Scholar]
- Canoll PD, Musacchio JM, Hardy R, Reynolds R, Marchionni MA, Salzer JL. GGF/neuregulin is a neuronal signal that promotes the proliferation and survival and inhibits the differentiation of oligodendrocyte progenitors. Neuron. 1996;17:229–243. doi: 10.1016/s0896-6273(00)80155-5. [DOI] [PubMed] [Google Scholar]
- Crews ST. Drosophila bHLH-PAS developmental regulatory proteins. In: Crews ST, editor. PAS Proteins: Regulators and Sensors of Development and Physiology. Kluwer; Boston: 2003. pp. 69–108. [Google Scholar]
- Crews ST. Axon-glial interactions at the Drosophila CNS midline. Cell Adh Migr. 2009;4:66–70. doi: 10.4161/cam.4.1.10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessaud E, McMahon AP, Briscoe J. Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development. 2008;135:2489–2503. doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- Dong R, Jacobs JR. Origin and differentiation of supernumerary midline glia in Drosophila embryos deficient for apoptosis. Dev Biol. 1997;190:165–177. doi: 10.1006/dbio.1997.8688. [DOI] [PubMed] [Google Scholar]
- Estes P, Fulkerson E, Zhang Y. Identification of motifs that are conserved in 12 Drosophila species and regulate midline glia vs. neuron expression. Genetics. 2008;178:787–799. doi: 10.1534/genetics.107.080440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol. 2010;11:276–287. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe VE. Mitotic domains reveal early commitment of cells in Drosophila embryos. Development. 1989;107:1–22. [PubMed] [Google Scholar]
- Fulkerson E, Estes PA. Common motifs shared by conserved enhancers of Drosophila midline glial genes. J Exp Zool B Mol Dev Evol. 2010;316:61–75. doi: 10.1002/jez.b.21382. [DOI] [PubMed] [Google Scholar]
- Garbe DS, Bashaw GJ. Axon guidance at the midline: from mutants to mechanisms. Crit Rev Biochem Mol Biol. 2004;39:319–341. doi: 10.1080/10409230490906797. [DOI] [PubMed] [Google Scholar]
- Giesen K, Lammel U, Langehans D, Krukkert K, Bunse I, Klämbt C. Regulation of glial cell number and differentiation by ecdysone and Fos signaling. Mech Dev. 2003;120:401–13. doi: 10.1016/s0925-4773(03)00009-1. [DOI] [PubMed] [Google Scholar]
- Go MJ, Eastman DS, Artavanis-Tsakonas S. Cell proliferation control by Notch signaling in Drosophila development. Development. 1998;125:2031–2040. doi: 10.1242/dev.125.11.2031. [DOI] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney JB, Wheeler SR, Estes P, Parente B, Crews ST. Gene expression profiling of the developing Drosophila CNS midline cells. Dev Biol. 2004;275:473–92. doi: 10.1016/j.ydbio.2004.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klämbt C, Jacobs JR, Goodman CS. The midline of the Drosophila central nervous system: a model for the genetic analysis of cell fate, cell migration, and growth cone guidance. Cell. 1991;64:801–815. doi: 10.1016/0092-8674(91)90509-w. [DOI] [PubMed] [Google Scholar]
- Lanoue BR, Gordon MD, Battye R, Jacobs JR. Genetic analysis of vein function in the Drosophila embryonic nervous system. Genome. 2000;43:564–573. [PubMed] [Google Scholar]
- Learte AR, Forero MG, Hidalgo A. Gliatrophic and gliatropic roles of PVF/PVR signaling during axon guidance. Glia. 2008;56:164–176. doi: 10.1002/glia.20601. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Maróy P, Kaufmann G, Dübendorfer A. Embryonic ecdysteroids of Drosophila melanogaster. J Insect Physiol. 1988;34:633–637. [Google Scholar]
- Noordermeer JN, Kopczynski CC, Fetter RD, Bland KS, Chen WY, Goodman CS. Wrapper, a novel member of the Ig superfamily, is expressed by midline glia and is required for them to ensheath commissural axons in Drosophila. 1998;21:991–1001. doi: 10.1016/s0896-6273(00)80618-2. [DOI] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, Mungall C, Svirskas R, Kadonaga JT, Doe CQ, Eisen MB, Celniker SE, Rubin GM. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci USA. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnepp B, Grumbling G, Donaldson T, Simcox A. Vein is a novel component in the Drosophila epidermal growth factor receptor pathway with similarity to the neuregulins. Genes Dev. 1996;10:2302–2313. doi: 10.1101/gad.10.18.2302. [DOI] [PubMed] [Google Scholar]
- Slovakova J, Carmena A. Canoe functions at the CNS midline glia in a complex with Shotgun and Wrapper-Nrx-IV during neuron-glia interactions. Development. 2011;138:1563–1571. doi: 10.1242/dev.056192. [DOI] [PubMed] [Google Scholar]
- Sonnenfeld MJ, Jacobs JR. Apoptosis of the midline glia during Drosophila embryogenesis: a correlation with axon contact. Development. 1995;121:569–578. doi: 10.1242/dev.121.2.569. [DOI] [PubMed] [Google Scholar]
- Stork T, Thomas S, Rodrigues F, Silies M, Naffin E, Wenderdel S, Klämbt C. Drosophila Neurexin IV stabilizes neuron-glia interactions at the CNS midline by binding to Wrapper. Development. 2009;136:1251–1261. doi: 10.1242/dev.032847. [DOI] [PubMed] [Google Scholar]
- Vasenkova I, Luginbuhl D, Chiba A. Gliopodia extend the range of direct glia-neuron communication during the CNS development in Drosophila. Mol Cell Neurosci. 2006;31:123–130. doi: 10.1016/j.mcn.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Verkhusha VV, Tsukita S, Oda H. Actin dynamics in lamellipodia of migrating border cells in the Drosophila ovary revealed by a GFP-actin fusion protein. FEBS Lett. 1999;445:395–401. doi: 10.1016/s0014-5793(99)00124-6. [DOI] [PubMed] [Google Scholar]
- Ward MP, Mosher JT, Crews ST. Regulation of bHLH-PAS protein subcellular localization during Drosophila embryogenesis. Development. 1998;125:1599–1608. doi: 10.1242/dev.125.9.1599. [DOI] [PubMed] [Google Scholar]
- Watson JD, Wheeler SR, Stagg SB, Crews ST. Drosophila hedgehog signaling and engrailed/runt mutual repression direct midline glia to alternative ensheathing and non-ensheathing fates. Development. 2011;138:1285–1295. doi: 10.1242/dev.056895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton JKA, Crews ST. CNS midline enhancers of the Drosophila slit and Toll genes. Mech Dev. 1993;40:141–154. doi: 10.1016/0925-4773(93)90072-6. [DOI] [PubMed] [Google Scholar]
- Wheeler SR, Kearney JB, Guardiola AR, Crews ST. Single-cell mapping of neural and glial gene expression in the developing Drosophila CNS midline cells. Dev Biol. 2006;294:509–524. doi: 10.1016/j.ydbio.2006.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler SR, Stagg SB, Crews ST. Multiple Notch signaling events control Drosophila CNS midline neurogenesis, gliogenesis and neuronal identity. Development. 2008;135:3071–3079. doi: 10.1242/dev.022343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler SR, Banerjee S, Blauth K, Rogers SL, Bhat MA, Crews ST. Neurexin IV and Wrapper interactions mediate Drosophila midline glial migration and axonal ensheathment. Development. 2009;136:1147–1157. doi: 10.1242/dev.030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka T, Tanaka O. Ultrastructural and cytochemical characterisation of the floor plate ependyma of the developing rat spinal cord. J Anat. 1989;165:87–100. [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Li B, Zhao W, Chang YH, Ma W, Dragan M, Barker JL, Hu Q, Rubinow DR. Sex-related differences in MAPKs activation in rat astrocytes: effects of estrogen on cell death. Brain Res Mol Brain Res. 2002;103:1–11. [PubMed] [Google Scholar]
- Zhou L, Schnitzler A, Agapite J, Schwartz LM, Steller H, Nambu JR. Cooperative functions of the reaper and head involution defective genes in the programmed cell death of Drosophila central nervous system midline cells. Proc Natl Acad Sci USA. 1997;94:5131–5136. doi: 10.1073/pnas.94.10.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Pennack JA, McQuilton P, Forero MG, Mizuguchi K, Sutcliffe B, Gu CJ, Fenton JC, Hidalgo A. Drosophila neurotrophins reveal a common mechanism for nervous system formation. PLoS Biol. 2008;6:2476–2495. doi: 10.1371/journal.pbio.0060284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A wave of apoptosis reduces AMG number
Time lapse imaging of a single segment from a glec-O1.5-Gal4 UAS-mCD8-GFP embryo in sagittal view. Anterior is to the left and dorsal is up. Movie corresponds to 1 hour and 14 minutes of development beginning during stage 13. In this segment, there are 11 AMG, not all AMG are in the focal plane. The AMG that are contacting the AC (ac) survive while the remainder fragment into apoptotic bodies and are cleared. Four individual AMG are indicated with colored arrows.
AMG migration to the DA region
Time lapse imaging of a single segment of a glec-O1.5-Gal4 UAS-mCD8-GFP embryo in sagittal view. Anterior is to the left and dorsal is up. Movie corresponds to 1 hour and 18 minutes of development beginning during stage 13 when only a single commissure (com) is present. Yellow arrow indicates the nucleus of an AMG that divides and then migrates dorsally to a position adjacent to the AC.
AMG migration to the VM region
Time lapse imaging of a single segment from a glec-O1.5-Gal4 UAS-mCD8-GFP embryo in sagittal view. Anterior is to the left and dorsal is up. Movie corresponds to 1 hour and 36 minutes of development beginning during stage 12/0. The single commissure is labeled “com”. Yellow arrow indicates the nucleus of an AMG that migrates dorsally to contact the AC.
AMG migration to the DM region
Time lapse imaging of a single segment from a glec-O1.5-Gal4 UAS-mCD8-GFP embryo in sagittal view. Anterior is to the left and dorsal is up. Movie corresponds to 32 minutes of development beginning during stage 14. Yellow arrow indicates the nucleus of an AMG that has paused below the AC (ac) before migrating between the AC and PC (pc). The AMG has a long process that extends posteriorly above the PC.
An individual AMG undergoes fragmentation and is cleared
Time lapse imaging of a single segment from a glec-O1.5-Gal4 UAS-mCD8-GFP embryo in sagittal view. Anterior is to the left and dorsal is up. Movie corresponds to 40 minutes of development beginning during stage 13. The AC and PC are labeled ac and pc. A single AMG (yellow arrow) that is in contact with the AC undergoes fragmentation and the debris is cleared.
PMG undergo fragmentation and are cleared
Time lapse imaging of a single segment from a glec-O1.5-Gal4 UAS-mCD8-GFP embryo in sagittal view. Anterior is to the left and dorsal is up. Movie corresponds to 1 hour and 8 minutes of development beginning during stage 14. The AC and PC are labeled “ac” and “pc”. A single PMG (yellow arrow) fragments and the debris is subsequently cleared. After fragmentation, cellular debris moves both dorsally (yellow arrow) and to the posterior (white arrow).
PMG nucleus extends towards and retracts from the PC
Time lapse imaging of a single segment from a glec-O1.5-Gal4 UAS-mCD8-GFP embryo in sagittal view. Anterior is to the left and dorsal is up. Movie corresponds to 52 minutes of development beginning during stage 16. The AC and PC are labeled “ac” and “pc”. PMG nucleus (yellow arrow) repeatedly extends towards and retracts from the PC.