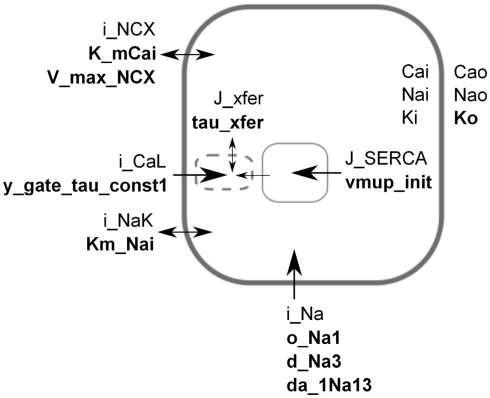
Figure 1.
Simplified schematic of the LNCS mouse heart cell model. For the sake of illustration, each parameter in the model was assumed to have a monogenic basis, with parameter values for genotypes aa, Aa, AA having parameter values of 50, 100, and 150% of baseline. Based on an initial sensitivity analysis, the parameters shown (bold) were selected for a full factorial simulation experiment representing 310 genotypes and their resulting phenotypes. The transmembrane potential is created by the difference in ion concentrations inside and outside the cell. Extracellular [Ca2+], [Na+], and [K+] are assumed constant, whereas the intracellular concentrations are state variables. Calcium is initially sequestered (J_SERCA) into the sarcoplasmic reticulum (SR, solid box) inside the cell. In response to an electric stimulus, the fast sodium current (i_Na) sets off an action potential that in turn triggers the release of calcium (J_xfer) via “dyadic space” microdomains (dashed box) where an early L-type calcium current (i_CaL) induces release of calcium from the SR. Parameters were selected to span a range of components and roles, see Table 1. Some components are strongly simplified in the figure, see Bondarenko et al. (2004), Li et al. (2010) for full description.