Abstract
In the crystal structure of the title complex, [Ni(C6H7NO)3](PF6)(C2F3O2), the NiII ion is in a slightly distorted octahedral NiO3N3 coordination geometry with each of the three N and three O atoms in a meridional coordination. In the crystal, the complex molecules and the trifluoroacetate anions are connected via O—H⋯O hydrogen bonding into layers parallel to the ab plane.
Related literature
For related complexes, see: Ito & Onaka (2004 ▶); Kermagoret & Braunstein (2008 ▶).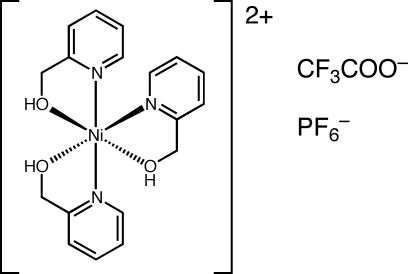
Experimental
Crystal data
[Ni(C6H7NO)3](PF6)(C2F3O2)
M r = 644.08
Triclinic,

a = 9.6381 (2) Å
b = 11.9668 (4) Å
c = 11.9892 (3) Å
α = 109.950 (1)°
β = 95.348 (1)°
γ = 101.411 (1)°
V = 1254.60 (6) Å3
Z = 2
Mo Kα radiation
μ = 0.94 mm−1
T = 200 K
0.40 × 0.30 × 0.20 mm
Data collection
Rigaku R-AXIS RAPID diffractometer
Absorption correction: multi-scan (ABSCOR; Rigaku, 1995 ▶) T min = 0.813, T max = 1.000
12517 measured reflections
5736 independent reflections
5234 reflections with I > 2σ(I)
R int = 0.018
Refinement
R[F 2 > 2σ(F 2)] = 0.033
wR(F 2) = 0.088
S = 1.05
5736 reflections
376 parameters
9 restraints
H-atom parameters constrained
Δρmax = 0.50 e Å−3
Δρmin = −0.45 e Å−3
Data collection: RAPID-AUTO (Rigaku, 2002 ▶); cell refinement: RAPID-AUTO; data reduction: RAPID-AUTO; program(s) used to solve structure: SIR2004 (Burla et al., 2005 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: Yadokari-XG (Wakita, 2001 ▶; Kabuto et al., 2009 ▶), ORTEP-3 for Windows (Farrugia, 1997 ▶) and Mercury (Macrae et al., 2008 ▶); software used to prepare material for publication: Yadokari-XG and publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S160053681104431X/nc2249sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S160053681104431X/nc2249Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Selected geometric parameters (Å, °).
| Ni1—O1 | 2.0461 (12) |
| Ni1—N2 | 2.0601 (14) |
| Ni1—O2 | 2.0647 (12) |
| Ni1—N1 | 2.0662 (14) |
| Ni1—O3 | 2.0714 (12) |
| Ni1—N3 | 2.0769 (14) |
| O1—Ni1—N1 | 78.11 (5) |
| N2—Ni1—O2 | 78.53 (5) |
| O3—Ni1—N3 | 78.09 (5) |
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯O4i | 0.87 | 1.76 | 2.6003 (19) | 162.5 |
| O2—H2⋯O5ii | 0.92 | 1.77 | 2.6965 (18) | 175.8 |
| O3—H3⋯O5iii | 0.98 | 1.65 | 2.6267 (18) | 173.8 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Acknowledgments
This work was supported in part by funds (No. 115001) from the Central Research Institute of Fukuoka University.
supplementary crystallographic information
Comment
The crystal structure of the title compound is composed of [NiII(C6H7ON)3]2+ cations, hexafluorophosphate and trifluoroacetate anions. The NiII ion is in a slightly distorted octahedral coordination, comprising three N atoms and three O atoms from three pyridine-2-methanol ligands (Fig. 1 and Table 1). The three N and three O atoms make a meridional NiO3N3 coordination and the mean bite angle of the pyridine-2-methanol ligand amount to 78.2 (2) °.
In the crystal structure the complexes are connected via O—H···O hydrogen bonding between the hydroxy H atoms of the pyridine-2-methanol ligand and the O atoms of the trifluoroacetate anion into layers that are parallel to the a/b plane. (Fig. 2 and 3 and Table 2).
Experimental
A solution of NiSO4.6H2O (0.5 mmol) in H2O (1 ml) was added to the solution of pyridine-2-methanol (1.5 mmol) in H2O (3 ml). Afterwards NH4PF6 (6.0 mmol) and CF3COONa (2.5 mmol) were added to the resulting blue solution. The resulting pale blue precipitate was collected. The crude product was purified by recrystallization from acetone and water. The blue prism-like crystals were obtained a few days later on slow evaporation of the solvent.
Refinement
The O–H H atoms were located in a difference Fourier map and the coordinates were fixed. Their Uiso(H) values were set to 1.5Ueq(O). Other H atoms were placed at calculated positions and were treated as riding on the parent C atoms, with C–H = 0.93 (CH) and 0.97 (CH2) Å and with Uiso(H) = 1.2Ueq(C). Three F atoms in CF3COO anions are rotationally disordered between three positions. The two parts of lower occupation were refined only isotropic (sof. 0.6:0.24:0.16).
Figures
Fig. 1.
ORTEP drawing for the title complex with labeling showing 50% probability displacement ellipsoids. Please note: The trifluoroacetate anion is disordered.
Fig. 2.
Crystal structure of the title compound view along the b-axis. The C-H H atoms, the PF6 anions and the disordered F atoms with lower occupation of the trifluoroacetate anions are omitted for clarity. O—H···O hydrogen bonding is shown as dashed blue lines.
Fig. 3.
Crystal structure of the title compound with view along the c-axis. The C-H H atoms and the disordered F atoms with lower occupation of the trifluoroacetate anions are omitted for clarity.
Crystal data
| [Ni(C6H7NO)3](PF6)(C2F3O2) | V = 1254.60 (6) Å3 |
| Mr = 644.08 | Z = 2 |
| Triclinic, P1 | F(000) = 652 |
| Hall symbol: -P 1 | Dx = 1.705 Mg m−3 |
| a = 9.6381 (2) Å | Mo Kα radiation, λ = 0.71075 Å |
| b = 11.9668 (4) Å | µ = 0.94 mm−1 |
| c = 11.9892 (3) Å | T = 200 K |
| α = 109.950 (1)° | Block, blue |
| β = 95.348 (1)° | 0.40 × 0.30 × 0.20 mm |
| γ = 101.411 (1)° |
Data collection
| Rigaku R-AXIS RAPID diffractometer | 5234 reflections with I > 2σ(I) |
| graphite | Rint = 0.018 |
| ω scans | θmax = 27.5°, θmin = 3.1° |
| Absorption correction: multi-scan (ABSCOR; Rigaku, 1995) | h = −12→11 |
| Tmin = 0.813, Tmax = 1.000 | k = −15→15 |
| 12517 measured reflections | l = −15→15 |
| 5736 independent reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.033 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.088 | H-atom parameters constrained |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0486P)2 + 0.5841P] where P = (Fo2 + 2Fc2)/3 |
| 5736 reflections | (Δ/σ)max = 0.001 |
| 376 parameters | Δρmax = 0.50 e Å−3 |
| 9 restraints | Δρmin = −0.45 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| Ni1 | 0.18393 (2) | 0.369025 (18) | 0.808996 (17) | 0.02212 (7) | |
| O1 | 0.00845 (13) | 0.39356 (12) | 0.88953 (12) | 0.0304 (3) | |
| H1 | −0.0596 | 0.4198 | 0.8609 | 0.046* | |
| O2 | 0.35252 (13) | 0.32008 (12) | 0.72559 (12) | 0.0337 (3) | |
| H2 | 0.4425 | 0.3733 | 0.7547 | 0.051* | |
| O3 | 0.33165 (14) | 0.49798 (11) | 0.95482 (11) | 0.0316 (3) | |
| H3 | 0.3431 | 0.5104 | 1.0408 | 0.047* | |
| N1 | 0.16982 (15) | 0.24566 (12) | 0.89561 (13) | 0.0257 (3) | |
| N2 | 0.07694 (15) | 0.24685 (13) | 0.64160 (13) | 0.0264 (3) | |
| N3 | 0.20297 (15) | 0.52595 (13) | 0.76794 (12) | 0.0255 (3) | |
| C1 | 0.26677 (19) | 0.17914 (16) | 0.90061 (18) | 0.0338 (4) | |
| H4 | 0.3381 | 0.1775 | 0.8509 | 0.041* | |
| C2 | 0.2656 (2) | 0.11372 (19) | 0.9755 (2) | 0.0447 (5) | |
| H5 | 0.3343 | 0.0670 | 0.9765 | 0.054* | |
| C3 | 0.1636 (3) | 0.1169 (2) | 1.0488 (2) | 0.0530 (6) | |
| H6 | 0.1624 | 0.0740 | 1.1025 | 0.064* | |
| C4 | 0.0627 (2) | 0.1836 (2) | 1.0432 (2) | 0.0458 (5) | |
| H7 | −0.0093 | 0.1864 | 1.0924 | 0.055* | |
| C5 | 0.06813 (18) | 0.24611 (15) | 0.96467 (16) | 0.0286 (3) | |
| C6 | −0.04137 (18) | 0.31692 (16) | 0.95294 (17) | 0.0297 (4) | |
| H8 | −0.1347 | 0.2596 | 0.9086 | 0.036* | |
| H9 | −0.0553 | 0.3674 | 1.0338 | 0.036* | |
| C7 | −0.06644 (19) | 0.20268 (16) | 0.60926 (16) | 0.0306 (4) | |
| H10 | −0.1254 | 0.2263 | 0.6681 | 0.037* | |
| C8 | −0.1309 (2) | 0.12438 (17) | 0.49383 (18) | 0.0377 (4) | |
| H11 | −0.2324 | 0.0945 | 0.4734 | 0.045* | |
| C9 | −0.0448 (2) | 0.09037 (18) | 0.40852 (18) | 0.0418 (5) | |
| H12 | −0.0866 | 0.0372 | 0.3282 | 0.050* | |
| C10 | 0.1021 (2) | 0.13424 (19) | 0.44087 (17) | 0.0400 (4) | |
| H13 | 0.1628 | 0.1117 | 0.3833 | 0.048* | |
| C11 | 0.1602 (2) | 0.21193 (17) | 0.55887 (16) | 0.0317 (4) | |
| C12 | 0.3203 (2) | 0.2616 (2) | 0.59846 (19) | 0.0488 (6) | |
| H14 | 0.3690 | 0.1939 | 0.5724 | 0.059* | |
| H15 | 0.3558 | 0.3211 | 0.5607 | 0.059* | |
| C13 | 0.1246 (2) | 0.53539 (18) | 0.67400 (16) | 0.0345 (4) | |
| H16 | 0.0528 | 0.4656 | 0.6217 | 0.041* | |
| C14 | 0.1440 (2) | 0.6415 (2) | 0.6504 (2) | 0.0446 (5) | |
| H17 | 0.0860 | 0.6451 | 0.5838 | 0.054* | |
| C15 | 0.2493 (2) | 0.7428 (2) | 0.7254 (2) | 0.0459 (5) | |
| H18 | 0.2664 | 0.8168 | 0.7102 | 0.055* | |
| C16 | 0.3291 (2) | 0.73469 (18) | 0.82255 (19) | 0.0375 (4) | |
| H19 | 0.4015 | 0.8034 | 0.8757 | 0.045* | |
| C17 | 0.30264 (17) | 0.62549 (15) | 0.84201 (15) | 0.0255 (3) | |
| C18 | 0.38677 (18) | 0.61428 (15) | 0.94787 (16) | 0.0282 (3) | |
| H20 | 0.4892 | 0.6238 | 0.9393 | 0.034* | |
| H21 | 0.3805 | 0.6801 | 1.0230 | 0.034* | |
| P1 | 0.59552 (5) | 0.02974 (4) | 0.74358 (4) | 0.03228 (11) | |
| F4 | 0.5841 (2) | 0.0626 (2) | 0.88218 (15) | 0.0891 (6) | |
| F5 | 0.61092 (18) | 0.00030 (18) | 0.60798 (13) | 0.0725 (5) | |
| F6 | 0.76632 (13) | 0.04945 (12) | 0.77770 (13) | 0.0494 (3) | |
| F7 | 0.61897 (16) | 0.17125 (12) | 0.76545 (17) | 0.0680 (5) | |
| F8 | 0.42616 (14) | 0.01136 (14) | 0.71068 (15) | 0.0597 (4) | |
| F9 | 0.57607 (18) | −0.10966 (14) | 0.7258 (2) | 0.0789 (5) | |
| F1A | 0.2162 (3) | 0.4114 (5) | 0.3777 (4) | 0.0633 (9) | 0.60 |
| F2A | 0.4293 (6) | 0.4201 (7) | 0.3364 (5) | 0.0959 (18) | 0.60 |
| F3A | 0.3715 (5) | 0.5861 (4) | 0.4479 (2) | 0.0956 (15) | 0.60 |
| F1B | 0.2326 (11) | 0.3584 (9) | 0.3289 (9) | 0.075 (3)* | 0.24 |
| F2B | 0.4556 (11) | 0.4729 (9) | 0.3644 (10) | 0.048 (2)* | 0.24 |
| F3B | 0.3095 (15) | 0.5496 (11) | 0.4460 (11) | 0.097 (4)* | 0.24 |
| F1C | 0.3378 (19) | 0.3456 (13) | 0.2889 (13) | 0.091 (4)* | 0.16 |
| F2C | 0.4376 (13) | 0.5252 (12) | 0.4082 (13) | 0.063 (3)* | 0.16 |
| F3C | 0.2240 (13) | 0.4706 (12) | 0.4110 (11) | 0.054 (3)* | 0.16 |
| O4 | 0.15453 (15) | 0.51056 (17) | 0.21720 (16) | 0.0494 (4) | |
| O5 | 0.38100 (14) | 0.53211 (14) | 0.18470 (12) | 0.0372 (3) | |
| C19 | 0.28069 (19) | 0.50937 (18) | 0.23870 (16) | 0.0320 (4) | |
| C20 | 0.3227 (3) | 0.4746 (3) | 0.3470 (2) | 0.0605 (7) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Ni1 | 0.01984 (11) | 0.02370 (12) | 0.02007 (11) | 0.00408 (8) | 0.00167 (7) | 0.00581 (8) |
| O1 | 0.0279 (6) | 0.0369 (7) | 0.0343 (7) | 0.0153 (5) | 0.0108 (5) | 0.0175 (6) |
| O2 | 0.0225 (6) | 0.0426 (7) | 0.0292 (6) | 0.0059 (5) | 0.0058 (5) | 0.0058 (5) |
| O3 | 0.0354 (7) | 0.0307 (6) | 0.0223 (6) | −0.0020 (5) | −0.0054 (5) | 0.0102 (5) |
| N1 | 0.0229 (6) | 0.0228 (6) | 0.0273 (7) | 0.0037 (5) | −0.0003 (5) | 0.0064 (5) |
| N2 | 0.0275 (7) | 0.0248 (7) | 0.0227 (7) | 0.0042 (5) | 0.0015 (5) | 0.0055 (5) |
| N3 | 0.0244 (7) | 0.0279 (7) | 0.0217 (7) | 0.0032 (5) | 0.0020 (5) | 0.0085 (5) |
| C1 | 0.0264 (8) | 0.0289 (9) | 0.0426 (10) | 0.0080 (7) | 0.0000 (7) | 0.0099 (8) |
| C2 | 0.0370 (10) | 0.0365 (10) | 0.0634 (14) | 0.0103 (8) | −0.0038 (9) | 0.0243 (10) |
| C3 | 0.0543 (13) | 0.0514 (13) | 0.0652 (15) | 0.0095 (10) | 0.0025 (11) | 0.0402 (12) |
| C4 | 0.0453 (12) | 0.0497 (12) | 0.0522 (13) | 0.0087 (9) | 0.0137 (10) | 0.0313 (11) |
| C5 | 0.0263 (8) | 0.0252 (8) | 0.0304 (8) | 0.0019 (6) | 0.0023 (6) | 0.0085 (7) |
| C6 | 0.0247 (8) | 0.0284 (8) | 0.0344 (9) | 0.0035 (6) | 0.0079 (7) | 0.0105 (7) |
| C7 | 0.0280 (8) | 0.0312 (9) | 0.0293 (9) | 0.0016 (7) | −0.0013 (7) | 0.0117 (7) |
| C8 | 0.0383 (10) | 0.0316 (9) | 0.0357 (10) | −0.0020 (7) | −0.0101 (8) | 0.0131 (8) |
| C9 | 0.0574 (13) | 0.0314 (9) | 0.0267 (9) | 0.0082 (9) | −0.0084 (8) | 0.0040 (8) |
| C10 | 0.0544 (12) | 0.0372 (10) | 0.0248 (9) | 0.0169 (9) | 0.0052 (8) | 0.0044 (8) |
| C11 | 0.0346 (9) | 0.0300 (9) | 0.0269 (8) | 0.0094 (7) | 0.0046 (7) | 0.0051 (7) |
| C12 | 0.0325 (10) | 0.0682 (15) | 0.0309 (10) | 0.0091 (9) | 0.0099 (8) | 0.0007 (10) |
| C13 | 0.0352 (9) | 0.0379 (10) | 0.0258 (8) | 0.0003 (7) | −0.0039 (7) | 0.0132 (7) |
| C14 | 0.0460 (11) | 0.0492 (12) | 0.0394 (11) | 0.0012 (9) | −0.0067 (9) | 0.0269 (10) |
| C15 | 0.0483 (12) | 0.0403 (11) | 0.0511 (13) | 0.0000 (9) | −0.0021 (10) | 0.0282 (10) |
| C16 | 0.0339 (9) | 0.0326 (9) | 0.0414 (10) | −0.0022 (7) | −0.0023 (8) | 0.0159 (8) |
| C17 | 0.0210 (7) | 0.0298 (8) | 0.0247 (8) | 0.0049 (6) | 0.0050 (6) | 0.0092 (7) |
| C18 | 0.0257 (8) | 0.0256 (8) | 0.0283 (8) | 0.0018 (6) | −0.0021 (6) | 0.0080 (7) |
| P1 | 0.0319 (2) | 0.0294 (2) | 0.0314 (2) | 0.00634 (18) | 0.00592 (18) | 0.00648 (19) |
| F4 | 0.0957 (13) | 0.1473 (19) | 0.0426 (9) | 0.0587 (13) | 0.0304 (9) | 0.0368 (11) |
| F5 | 0.0719 (10) | 0.1113 (14) | 0.0344 (7) | 0.0355 (10) | 0.0129 (7) | 0.0187 (8) |
| F6 | 0.0357 (6) | 0.0454 (7) | 0.0592 (8) | 0.0106 (5) | 0.0001 (5) | 0.0114 (6) |
| F7 | 0.0577 (9) | 0.0331 (7) | 0.1023 (13) | 0.0123 (6) | −0.0049 (8) | 0.0157 (8) |
| F8 | 0.0328 (7) | 0.0575 (8) | 0.0740 (10) | 0.0086 (6) | 0.0046 (6) | 0.0086 (7) |
| F9 | 0.0600 (9) | 0.0381 (7) | 0.1392 (17) | 0.0055 (7) | 0.0159 (10) | 0.0374 (9) |
| F1A | 0.0651 (18) | 0.089 (3) | 0.061 (2) | 0.0150 (19) | 0.0181 (15) | 0.059 (2) |
| F2A | 0.082 (3) | 0.167 (5) | 0.110 (4) | 0.085 (4) | 0.032 (3) | 0.105 (4) |
| F3A | 0.075 (2) | 0.159 (4) | 0.0224 (12) | −0.018 (3) | −0.0068 (13) | 0.0261 (17) |
| O4 | 0.0281 (7) | 0.0754 (11) | 0.0610 (10) | 0.0183 (7) | 0.0077 (6) | 0.0425 (9) |
| O5 | 0.0281 (6) | 0.0558 (8) | 0.0263 (6) | 0.0041 (6) | 0.0028 (5) | 0.0171 (6) |
| C19 | 0.0276 (8) | 0.0416 (10) | 0.0291 (9) | 0.0093 (7) | 0.0032 (7) | 0.0158 (8) |
| C20 | 0.0362 (11) | 0.114 (2) | 0.0521 (14) | 0.0214 (13) | 0.0112 (10) | 0.0533 (16) |
Geometric parameters (Å, °)
| Ni1—O1 | 2.0461 (12) | C10—C11 | 1.389 (3) |
| Ni1—N2 | 2.0601 (14) | C10—H13 | 0.9500 |
| Ni1—O2 | 2.0647 (12) | C11—C12 | 1.506 (3) |
| Ni1—N1 | 2.0662 (14) | C12—H14 | 0.9900 |
| Ni1—O3 | 2.0714 (12) | C12—H15 | 0.9900 |
| Ni1—N3 | 2.0769 (14) | C13—C14 | 1.374 (3) |
| O1—C6 | 1.421 (2) | C13—H16 | 0.9500 |
| O1—H1 | 0.8716 | C14—C15 | 1.381 (3) |
| O2—C12 | 1.417 (2) | C14—H17 | 0.9500 |
| O2—H2 | 0.9235 | C15—C16 | 1.377 (3) |
| O3—C18 | 1.422 (2) | C15—H18 | 0.9500 |
| O3—H3 | 0.9822 | C16—C17 | 1.384 (3) |
| N1—C5 | 1.340 (2) | C16—H19 | 0.9500 |
| N1—C1 | 1.352 (2) | C17—C18 | 1.501 (2) |
| N2—C11 | 1.342 (2) | C18—H20 | 0.9900 |
| N2—C7 | 1.346 (2) | C18—H21 | 0.9900 |
| N3—C17 | 1.340 (2) | P1—F5 | 1.5697 (15) |
| N3—C13 | 1.347 (2) | P1—F9 | 1.5777 (15) |
| C1—C2 | 1.377 (3) | P1—F7 | 1.5881 (14) |
| C1—H4 | 0.9500 | P1—F4 | 1.5915 (16) |
| C2—C3 | 1.376 (4) | P1—F8 | 1.5960 (14) |
| C2—H5 | 0.9500 | P1—F6 | 1.6087 (13) |
| C3—C4 | 1.385 (3) | F1A—C20 | 1.308 (4) |
| C3—H6 | 0.9500 | F2A—C20 | 1.316 (4) |
| C4—C5 | 1.387 (3) | F3A—C20 | 1.417 (5) |
| C4—H7 | 0.9500 | F1B—C20 | 1.420 (10) |
| C5—C6 | 1.503 (2) | F2B—C20 | 1.284 (11) |
| C6—H8 | 0.9900 | F3B—C20 | 1.255 (12) |
| C6—H9 | 0.9900 | F1C—C20 | 1.505 (14) |
| C7—C8 | 1.380 (3) | F2C—C20 | 1.188 (11) |
| C7—H10 | 0.9500 | F3C—C20 | 1.279 (12) |
| C8—C9 | 1.381 (3) | O4—C19 | 1.223 (2) |
| C8—H11 | 0.9500 | O5—C19 | 1.249 (2) |
| C9—C10 | 1.376 (3) | C19—C20 | 1.536 (3) |
| C9—H12 | 0.9500 | ||
| O1—Ni1—N2 | 98.09 (6) | N2—C11—C10 | 121.78 (18) |
| O1—Ni1—O2 | 172.33 (5) | N2—C11—C12 | 117.35 (16) |
| O1—Ni1—N1 | 78.11 (5) | C10—C11—C12 | 120.87 (18) |
| N2—Ni1—O2 | 78.53 (5) | O2—C12—C11 | 109.50 (16) |
| N2—Ni1—N1 | 97.35 (6) | O2—C12—H14 | 109.8 |
| O2—Ni1—N1 | 95.41 (6) | C11—C12—H14 | 109.8 |
| O1—Ni1—O3 | 95.00 (5) | O2—C12—H15 | 109.8 |
| N2—Ni1—O3 | 166.15 (6) | C11—C12—H15 | 109.8 |
| O2—Ni1—O3 | 89.01 (5) | H14—C12—H15 | 108.2 |
| N1—Ni1—O3 | 89.74 (5) | N3—C13—C14 | 122.91 (17) |
| O1—Ni1—N3 | 93.77 (5) | N3—C13—H16 | 118.5 |
| N2—Ni1—N3 | 96.49 (6) | C14—C13—H16 | 118.5 |
| O2—Ni1—N3 | 93.45 (6) | C13—C14—C15 | 118.77 (18) |
| N1—Ni1—N3 | 164.81 (6) | C13—C14—H17 | 120.6 |
| O3—Ni1—N3 | 78.09 (5) | C15—C14—H17 | 120.6 |
| C6—O1—Ni1 | 118.49 (10) | C16—C15—C14 | 118.86 (18) |
| C6—O1—H1 | 112.8 | C16—C15—H18 | 120.6 |
| Ni1—O1—H1 | 122.2 | C14—C15—H18 | 120.6 |
| C12—O2—Ni1 | 115.90 (11) | C15—C16—C17 | 119.37 (18) |
| C12—O2—H2 | 114.4 | C15—C16—H19 | 120.3 |
| Ni1—O2—H2 | 118.1 | C17—C16—H19 | 120.3 |
| C18—O3—Ni1 | 117.12 (10) | N3—C17—C16 | 122.05 (16) |
| C18—O3—H3 | 106.7 | N3—C17—C18 | 117.36 (15) |
| Ni1—O3—H3 | 130.7 | C16—C17—C18 | 120.58 (15) |
| C5—N1—C1 | 118.61 (16) | O3—C18—C17 | 110.11 (13) |
| C5—N1—Ni1 | 116.13 (11) | O3—C18—H20 | 109.6 |
| C1—N1—Ni1 | 124.57 (13) | C17—C18—H20 | 109.6 |
| C11—N2—C7 | 118.69 (15) | O3—C18—H21 | 109.6 |
| C11—N2—Ni1 | 115.61 (12) | C17—C18—H21 | 109.6 |
| C7—N2—Ni1 | 125.69 (12) | H20—C18—H21 | 108.2 |
| C17—N3—C13 | 118.01 (15) | F5—P1—F9 | 90.77 (11) |
| C17—N3—Ni1 | 116.35 (11) | F5—P1—F7 | 90.50 (11) |
| C13—N3—Ni1 | 125.64 (12) | F9—P1—F7 | 178.09 (10) |
| N1—C1—C2 | 122.16 (19) | F5—P1—F4 | 178.33 (12) |
| N1—C1—H4 | 118.9 | F9—P1—F4 | 90.26 (12) |
| C2—C1—H4 | 118.9 | F7—P1—F4 | 88.43 (12) |
| C3—C2—C1 | 119.20 (19) | F5—P1—F8 | 90.97 (9) |
| C3—C2—H5 | 120.4 | F9—P1—F8 | 91.58 (9) |
| C1—C2—H5 | 120.4 | F7—P1—F8 | 89.83 (8) |
| C2—C3—C4 | 119.1 (2) | F4—P1—F8 | 90.31 (10) |
| C2—C3—H6 | 120.5 | F5—P1—F6 | 89.37 (8) |
| C4—C3—H6 | 120.5 | F9—P1—F6 | 88.81 (8) |
| C3—C4—C5 | 119.1 (2) | F7—P1—F6 | 89.77 (7) |
| C3—C4—H7 | 120.5 | F4—P1—F6 | 89.35 (9) |
| C5—C4—H7 | 120.5 | F8—P1—F6 | 179.48 (9) |
| N1—C5—C4 | 121.87 (17) | O4—C19—O5 | 129.17 (18) |
| N1—C5—C6 | 117.09 (15) | O4—C19—C20 | 115.85 (17) |
| C4—C5—C6 | 121.03 (17) | O5—C19—C20 | 114.97 (16) |
| O1—C6—C5 | 108.67 (14) | F2C—C20—F3C | 111.5 (9) |
| O1—C6—H8 | 110.0 | F3B—C20—F2B | 102.8 (7) |
| C5—C6—H8 | 110.0 | F1A—C20—F2A | 109.8 (4) |
| O1—C6—H9 | 110.0 | F1A—C20—F3A | 104.4 (3) |
| C5—C6—H9 | 110.0 | F2A—C20—F3A | 106.6 (4) |
| H8—C6—H9 | 108.3 | F3B—C20—F1B | 108.1 (7) |
| N2—C7—C8 | 122.40 (18) | F2B—C20—F1B | 110.5 (6) |
| N2—C7—H10 | 118.8 | F2C—C20—F1C | 102.0 (9) |
| C8—C7—H10 | 118.8 | F3C—C20—F1C | 107.2 (8) |
| C7—C8—C9 | 118.65 (18) | F2C—C20—C19 | 118.3 (6) |
| C7—C8—H11 | 120.7 | F3B—C20—C19 | 113.8 (6) |
| C9—C8—H11 | 120.7 | F3C—C20—C19 | 113.1 (6) |
| C10—C9—C8 | 119.43 (18) | F2B—C20—C19 | 113.3 (5) |
| C10—C9—H12 | 120.3 | F1A—C20—C19 | 115.0 (2) |
| C8—C9—H12 | 120.3 | F2A—C20—C19 | 113.6 (3) |
| C9—C10—C11 | 119.03 (19) | F3A—C20—C19 | 106.5 (3) |
| C9—C10—H13 | 120.5 | F1B—C20—C19 | 108.2 (4) |
| C11—C10—H13 | 120.5 | F1C—C20—C19 | 103.1 (6) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···O4i | 0.87 | 1.76 | 2.6003 (19) | 162.5 |
| O2—H2···O5ii | 0.92 | 1.77 | 2.6965 (18) | 175.8 |
| O3—H3···O5iii | 0.98 | 1.65 | 2.6267 (18) | 173.8 |
Symmetry codes: (i) −x, −y+1, −z+1; (ii) −x+1, −y+1, −z+1; (iii) x, y, z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: NC2249).
References
- Burla, M. C., Caliandro, R., Camalli, M., Carrozzini, B., Cascarano, G. L., De Caro, L., Giacovazzo, C., Polidori, G. & Spagna, R. (2005). J. Appl. Cryst. 38, 381–388.
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Ito, M. & Onaka, S. (2004). Inorg. Chim. Acta, 357, 1039–1046.
- Kabuto, C., Akine, S., Nemoto, T. & Kwon, E. (2009). J. Cryst. Soc. Jpn, 51, 218–224.
- Kermagoret, A. & Braunstein, P. (2008). Dalton Trans. pp. 1564–1573. [DOI] [PubMed]
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Rigaku (1995). ABSCOR Rigaku Corporation, Tokyo, Japan.
- Rigaku (2002). RAPID-AUTO Rigaku Corporation, Tokyo, Japan.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Wakita, K. (2001). Yadokari-XG. http://www.hat.hi-ho.ne.jp/k-wakita/yadokari.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S160053681104431X/nc2249sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S160053681104431X/nc2249Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report





