Abstract
In the title compound, C25H15Cl2N, the benzo[h]quinoline system exhibits an approximately planar conformation with an r.m.s. deviation of 0.0202Å and a maximum deviation of 0.039 (1) Å. The aryl group at position 2 is nearly coplanar with the parent ring [dihedral angle = 6.68 (7)°] while the parent ring and the phenyl subsitituent at position 4 form a dihedral angle of 67.11 (4)°. Intermolecular C—H⋯π interactions stabilize the crystal packing.
Related literature
For the uses of metal complexes of benzo[h]quinoline as electronic materials and organic electronic devices, see: Cho et al. (2010 ▶). For the medicinal uses of benzo[h]quinoline and its complexes, see: Pantoom et al. (2011 ▶); Liu et al. (2011 ▶). For the preparation of the title compound, see: Zhang et al. (2010 ▶).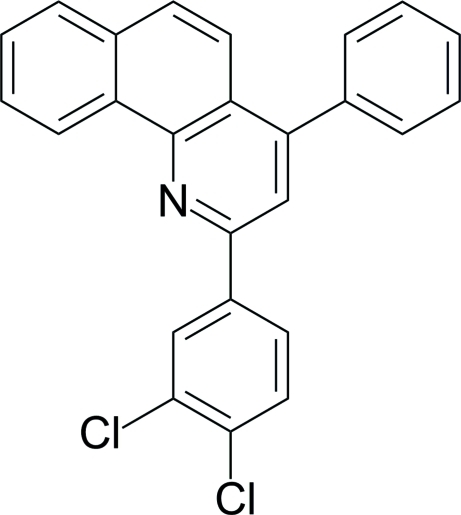
Experimental
Crystal data
C25H15Cl2N
M r = 400.28
Monoclinic,
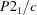
a = 10.6066 (14) Å
b = 9.5667 (12) Å
c = 18.824 (2) Å
β = 94.264 (7)°
V = 1904.8 (4) Å3
Z = 4
Mo Kα radiation
μ = 0.35 mm−1
T = 113 K
0.20 × 0.18 × 0.12 mm
Data collection
Rigaku Saturn724 CCD diffractometer
Absorption correction: multi-scan (CrystalClear; Rigaku/MSC, 2002) ▶ T min = 0.933, T max = 0.959
23687 measured reflections
4523 independent reflections
3630 reflections with I > 2σ(I)
R int = 0.045
Refinement
R[F 2 > 2σ(F 2)] = 0.039
wR(F 2) = 0.110
S = 1.07
4523 reflections
253 parameters
H-atom parameters constrained
Δρmax = 0.37 e Å−3
Δρmin = −0.36 e Å−3
Data collection: CrystalClear (Rigaku/MSC, 2002) ▶; cell refinement: CrystalClear ▶; data reduction: CrystalClear ▶; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536811040049/hg5101sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811040049/hg5101Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811040049/hg5101Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg1, Cg2 and Cg3 are the centroids of the C20–C25, C14–C19 and N1/C1/C10–C13 rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C6—H6⋯Cg1i | 0.95 | 2.98 | 3.8577 (19) | 154 |
| C22—H22⋯Cg2ii | 0.95 | 2.94 | 3.8204 (19) | 156 |
| C25—H25⋯Cg3iii | 0.95 | 2.63 | 3.4738 (17) | 148 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Acknowledgments
We are grateful to the Special Presidential Foundation of Xuzhou Medical College (2010KJZ15) for financial support.
supplementary crystallographic information
Comment
The benzo[h]quinoline derivatives and their complexes can be used as electronic material and organic electronic device (Cho et al. ,2010), potent family-18 chitinase inhibitors (Pantoom et al., 2011), topoisomerase IIα poisons (Liu et al., 2011). Besides, They can also treat Alzheimer's disease. These properties arouse our interset in the relationship between their structures and activities. During the synthesis of benzo[h]quinoline derivatives, the title compound, (I) was isolated and its structure was determined by X-ray diffraction. Herein we shall report its crystal structure. The molecular structure of (I) is shown in Fig. 1. In the molecular structure, the benzo[h]quinoline exhibits a planar conformation with RMS of 0.0202Å and the largest deviation is 0.039 (1) Å. The 3,4-dichlorophenyl is almost coplanar with benzo[h]quinoline, since the dihedral angle between them is only 6.68 (7)°. The parent ring and the phenyl subsitituent at position 4 form a dihedral angle of 67.11 (4)°. In addition, there is a non-classical intramolecular hydrogen bond (C19—H19···N1). The crystal packing is stabilized by the intermolecular C—H···π interactions (Fig. 2, Table 1).
Experimental
The title compound was synthesized according to the reported procedure (Zhang et al., 2010). Under an air atmosphere, a 10 ml of sealable reaction tube equipped with a magnetic stir bar was charged with an 3,4-dichlorobenzaldehyde (1.00 mmol), naphthalen-1-amine (1.00 mmol), and the mixture was heated and stirred in an oil bath at 333 K for 1 h. Then FeCl3 (16.2 mg, 0.10 mmol), ethynylbenzene (1.10 mmol) were added. The reaction mixture was then stirred in an oil bath at 393 K until the substrates were consumed completely (about 12 h), and then it was cooled to room temperature and the solvent was evaporated, the residue was purified by flash chromatography(hexane/AcOEt = 15:1) to afford the desired product.The single-crystal suitable for X-ray diffraction was obtained through the evaporation of ethanol solution.
Refinement
All H atoms were placed in calculated positions, with C—H = 0.95 Å, and included in the final cycles of refinement using a riding model, with Uĩso~(H) = 1.2U~eq~(parent atom).
Figures
Fig. 1.
The structure of (I), showing 30% probability displacement ellipsoids and the atom-numbering scheme. Cg1 is the centroid of the ring of C20/C21/C22/C23/C24/C25. Cg2 is the centroid of the ring of C14/C15/C16/C17/C18/C19. Cg3 is the centroid of the ring of N1/C1/C10/C11/C12/C13.
Fig. 2.
The packing diagram of (I).
Crystal data
| C25H15Cl2N | F(000) = 824 |
| Mr = 400.28 | Dx = 1.396 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 6694 reflections |
| a = 10.6066 (14) Å | θ = 1.9–27.9° |
| b = 9.5667 (12) Å | µ = 0.35 mm−1 |
| c = 18.824 (2) Å | T = 113 K |
| β = 94.264 (7)° | Prism, colorless |
| V = 1904.8 (4) Å3 | 0.20 × 0.18 × 0.12 mm |
| Z = 4 |
Data collection
| Rigaku Saturn724 CCD diffractometer | 4523 independent reflections |
| Radiation source: rotating anode | 3630 reflections with I > 2σ(I) |
| multilayer | Rint = 0.045 |
| Detector resolution: 14.22 pixels mm-1 | θmax = 27.8°, θmin = 1.9° |
| ω and φ scans | h = −13→13 |
| Absorption correction: multi-scan (CrystalClear; Rigaku/MSC, 2002) | k = −12→12 |
| Tmin = 0.933, Tmax = 0.959 | l = −24→24 |
| 23687 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.039 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.110 | H-atom parameters constrained |
| S = 1.07 | w = 1/[σ2(Fo2) + (0.0602P)2] where P = (Fo2 + 2Fc2)/3 |
| 4523 reflections | (Δ/σ)max = 0.002 |
| 253 parameters | Δρmax = 0.37 e Å−3 |
| 0 restraints | Δρmin = −0.36 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.52078 (4) | 0.42736 (4) | 1.09062 (2) | 0.03323 (13) | |
| Cl2 | 0.56082 (4) | 0.57108 (5) | 1.24136 (2) | 0.03876 (14) | |
| N1 | 0.09940 (11) | 0.64203 (12) | 0.98050 (6) | 0.0220 (3) | |
| C1 | −0.00408 (13) | 0.66067 (15) | 0.93439 (8) | 0.0217 (3) | |
| C2 | −0.00284 (14) | 0.59559 (15) | 0.86472 (8) | 0.0226 (3) | |
| C3 | 0.10040 (15) | 0.51461 (17) | 0.84545 (8) | 0.0270 (4) | |
| H3 | 0.1729 | 0.5043 | 0.8778 | 0.032* | |
| C4 | 0.09621 (16) | 0.45062 (16) | 0.78003 (9) | 0.0310 (4) | |
| H4 | 0.1656 | 0.3950 | 0.7678 | 0.037* | |
| C5 | −0.00939 (16) | 0.46621 (17) | 0.73071 (9) | 0.0318 (4) | |
| H5 | −0.0110 | 0.4210 | 0.6857 | 0.038* | |
| C6 | −0.11019 (16) | 0.54695 (16) | 0.74773 (9) | 0.0292 (4) | |
| H6 | −0.1807 | 0.5586 | 0.7141 | 0.035* | |
| C7 | −0.10939 (14) | 0.61255 (16) | 0.81487 (8) | 0.0239 (3) | |
| C8 | −0.21411 (14) | 0.69559 (16) | 0.83422 (8) | 0.0260 (3) | |
| H8 | −0.2849 | 0.7073 | 0.8008 | 0.031* | |
| C9 | −0.21506 (14) | 0.75764 (15) | 0.89867 (8) | 0.0264 (3) | |
| H9 | −0.2853 | 0.8136 | 0.9092 | 0.032* | |
| C10 | −0.11070 (13) | 0.73999 (15) | 0.95172 (8) | 0.0231 (3) | |
| C11 | −0.10774 (14) | 0.79943 (15) | 1.02076 (8) | 0.0236 (3) | |
| C12 | −0.00148 (14) | 0.78015 (16) | 1.06638 (8) | 0.0249 (3) | |
| H12 | 0.0027 | 0.8206 | 1.1126 | 0.030* | |
| C13 | 0.10125 (14) | 0.70060 (15) | 1.04485 (8) | 0.0229 (3) | |
| C14 | 0.21694 (14) | 0.67568 (15) | 1.09325 (8) | 0.0231 (3) | |
| C15 | 0.23628 (14) | 0.73938 (16) | 1.15973 (8) | 0.0291 (4) | |
| H15 | 0.1761 | 0.8048 | 1.1745 | 0.035* | |
| C16 | 0.34251 (14) | 0.70838 (17) | 1.20477 (8) | 0.0311 (4) | |
| H16 | 0.3546 | 0.7529 | 1.2499 | 0.037* | |
| C17 | 0.43053 (14) | 0.61299 (17) | 1.18414 (8) | 0.0276 (4) | |
| C18 | 0.41328 (14) | 0.55001 (15) | 1.11722 (8) | 0.0250 (3) | |
| C19 | 0.30761 (14) | 0.58159 (15) | 1.07252 (8) | 0.0238 (3) | |
| H19 | 0.2967 | 0.5385 | 1.0270 | 0.029* | |
| C20 | −0.21559 (14) | 0.88347 (16) | 1.04496 (8) | 0.0245 (3) | |
| C21 | −0.33198 (14) | 0.82296 (18) | 1.05638 (9) | 0.0328 (4) | |
| H21 | −0.3450 | 0.7260 | 1.0476 | 0.039* | |
| C22 | −0.42876 (16) | 0.90333 (19) | 1.08044 (9) | 0.0361 (4) | |
| H22 | −0.5078 | 0.8614 | 1.0881 | 0.043* | |
| C23 | −0.41049 (16) | 1.04477 (17) | 1.09328 (9) | 0.0335 (4) | |
| H23 | −0.4769 | 1.0996 | 1.1100 | 0.040* | |
| C24 | −0.29624 (16) | 1.10611 (18) | 1.08182 (9) | 0.0330 (4) | |
| H24 | −0.2842 | 1.2034 | 1.0899 | 0.040* | |
| C25 | −0.19865 (15) | 1.02549 (16) | 1.05835 (8) | 0.0274 (4) | |
| H25 | −0.1195 | 1.0678 | 1.0514 | 0.033* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0260 (2) | 0.0342 (2) | 0.0393 (3) | 0.01057 (17) | 0.00126 (18) | −0.00259 (18) |
| Cl2 | 0.0285 (2) | 0.0490 (3) | 0.0374 (3) | 0.00308 (19) | −0.00704 (18) | −0.0028 (2) |
| N1 | 0.0228 (7) | 0.0176 (6) | 0.0260 (7) | 0.0012 (5) | 0.0046 (5) | 0.0016 (5) |
| C1 | 0.0220 (8) | 0.0166 (7) | 0.0269 (8) | −0.0008 (6) | 0.0039 (6) | 0.0018 (6) |
| C2 | 0.0223 (8) | 0.0188 (7) | 0.0270 (8) | −0.0010 (6) | 0.0047 (6) | 0.0022 (6) |
| C3 | 0.0245 (8) | 0.0254 (8) | 0.0312 (9) | 0.0021 (7) | 0.0023 (7) | −0.0014 (7) |
| C4 | 0.0302 (9) | 0.0294 (9) | 0.0339 (9) | 0.0042 (7) | 0.0059 (7) | −0.0066 (7) |
| C5 | 0.0343 (9) | 0.0322 (9) | 0.0292 (9) | −0.0019 (8) | 0.0046 (7) | −0.0052 (7) |
| C6 | 0.0278 (9) | 0.0296 (9) | 0.0298 (9) | −0.0028 (7) | −0.0002 (7) | 0.0016 (7) |
| C7 | 0.0232 (8) | 0.0202 (8) | 0.0287 (8) | −0.0026 (6) | 0.0038 (6) | 0.0033 (6) |
| C8 | 0.0214 (8) | 0.0260 (8) | 0.0306 (8) | −0.0008 (6) | 0.0006 (6) | 0.0066 (7) |
| C9 | 0.0210 (7) | 0.0234 (8) | 0.0354 (9) | 0.0028 (6) | 0.0055 (7) | 0.0040 (7) |
| C10 | 0.0205 (7) | 0.0194 (7) | 0.0298 (8) | 0.0005 (6) | 0.0055 (6) | 0.0032 (6) |
| C11 | 0.0234 (8) | 0.0184 (7) | 0.0297 (8) | 0.0003 (6) | 0.0078 (6) | 0.0038 (6) |
| C12 | 0.0281 (8) | 0.0215 (8) | 0.0259 (8) | 0.0024 (6) | 0.0072 (6) | 0.0007 (6) |
| C13 | 0.0238 (8) | 0.0173 (7) | 0.0279 (8) | −0.0009 (6) | 0.0051 (6) | 0.0027 (6) |
| C14 | 0.0239 (8) | 0.0188 (7) | 0.0272 (8) | −0.0008 (6) | 0.0054 (6) | 0.0022 (6) |
| C15 | 0.0288 (9) | 0.0267 (8) | 0.0323 (9) | 0.0042 (7) | 0.0050 (7) | −0.0027 (7) |
| C16 | 0.0329 (9) | 0.0313 (9) | 0.0291 (9) | −0.0019 (7) | 0.0024 (7) | −0.0055 (7) |
| C17 | 0.0235 (8) | 0.0284 (9) | 0.0302 (9) | −0.0026 (7) | −0.0022 (7) | 0.0029 (7) |
| C18 | 0.0228 (8) | 0.0214 (8) | 0.0314 (9) | 0.0013 (6) | 0.0060 (7) | 0.0008 (7) |
| C19 | 0.0256 (8) | 0.0217 (8) | 0.0242 (8) | 0.0004 (6) | 0.0030 (6) | 0.0001 (6) |
| C20 | 0.0246 (8) | 0.0245 (8) | 0.0249 (8) | 0.0049 (6) | 0.0052 (6) | 0.0037 (6) |
| C21 | 0.0291 (9) | 0.0256 (9) | 0.0447 (10) | −0.0001 (7) | 0.0105 (8) | 0.0016 (7) |
| C22 | 0.0276 (9) | 0.0366 (10) | 0.0458 (11) | 0.0022 (8) | 0.0142 (8) | 0.0046 (8) |
| C23 | 0.0307 (9) | 0.0345 (10) | 0.0368 (10) | 0.0113 (8) | 0.0129 (7) | 0.0052 (8) |
| C24 | 0.0345 (9) | 0.0256 (9) | 0.0399 (10) | 0.0085 (7) | 0.0096 (8) | 0.0018 (7) |
| C25 | 0.0252 (8) | 0.0253 (8) | 0.0324 (9) | 0.0021 (7) | 0.0073 (7) | 0.0036 (7) |
Geometric parameters (Å, °)
| Cl1—C18 | 1.7356 (15) | C12—C13 | 1.413 (2) |
| Cl2—C17 | 1.7347 (16) | C12—H12 | 0.9500 |
| N1—C13 | 1.3334 (18) | C13—C14 | 1.492 (2) |
| N1—C1 | 1.3595 (19) | C14—C15 | 1.393 (2) |
| C1—C10 | 1.420 (2) | C14—C19 | 1.394 (2) |
| C1—C2 | 1.453 (2) | C15—C16 | 1.391 (2) |
| C2—C3 | 1.411 (2) | C15—H15 | 0.9500 |
| C2—C7 | 1.423 (2) | C16—C17 | 1.382 (2) |
| C3—C4 | 1.373 (2) | C16—H16 | 0.9500 |
| C3—H3 | 0.9500 | C17—C18 | 1.396 (2) |
| C4—C5 | 1.409 (2) | C18—C19 | 1.384 (2) |
| C4—H4 | 0.9500 | C19—H19 | 0.9500 |
| C5—C6 | 1.376 (2) | C20—C25 | 1.391 (2) |
| C5—H5 | 0.9500 | C20—C21 | 1.395 (2) |
| C6—C7 | 1.411 (2) | C21—C22 | 1.385 (2) |
| C6—H6 | 0.9500 | C21—H21 | 0.9500 |
| C7—C8 | 1.435 (2) | C22—C23 | 1.386 (2) |
| C8—C9 | 1.351 (2) | C22—H22 | 0.9500 |
| C8—H8 | 0.9500 | C23—C24 | 1.378 (2) |
| C9—C10 | 1.444 (2) | C23—H23 | 0.9500 |
| C9—H9 | 0.9500 | C24—C25 | 1.389 (2) |
| C10—C11 | 1.417 (2) | C24—H24 | 0.9500 |
| C11—C12 | 1.378 (2) | C25—H25 | 0.9500 |
| C11—C20 | 1.497 (2) | ||
| C13—N1—C1 | 118.79 (12) | N1—C13—C14 | 116.32 (13) |
| N1—C1—C10 | 122.84 (14) | C12—C13—C14 | 121.84 (14) |
| N1—C1—C2 | 117.34 (13) | C15—C14—C19 | 118.38 (14) |
| C10—C1—C2 | 119.82 (13) | C15—C14—C13 | 122.55 (14) |
| C3—C2—C7 | 119.10 (14) | C19—C14—C13 | 119.04 (14) |
| C3—C2—C1 | 121.79 (14) | C16—C15—C14 | 120.85 (14) |
| C7—C2—C1 | 119.10 (13) | C16—C15—H15 | 119.6 |
| C4—C3—C2 | 120.10 (14) | C14—C15—H15 | 119.6 |
| C4—C3—H3 | 119.9 | C17—C16—C15 | 120.21 (15) |
| C2—C3—H3 | 119.9 | C17—C16—H16 | 119.9 |
| C3—C4—C5 | 121.00 (15) | C15—C16—H16 | 119.9 |
| C3—C4—H4 | 119.5 | C16—C17—C18 | 119.52 (14) |
| C5—C4—H4 | 119.5 | C16—C17—Cl2 | 120.12 (12) |
| C6—C5—C4 | 119.97 (15) | C18—C17—Cl2 | 120.36 (12) |
| C6—C5—H5 | 120.0 | C19—C18—C17 | 120.01 (14) |
| C4—C5—H5 | 120.0 | C19—C18—Cl1 | 119.47 (12) |
| C5—C6—C7 | 120.35 (15) | C17—C18—Cl1 | 120.48 (12) |
| C5—C6—H6 | 119.8 | C18—C19—C14 | 121.01 (14) |
| C7—C6—H6 | 119.8 | C18—C19—H19 | 119.5 |
| C6—C7—C2 | 119.45 (14) | C14—C19—H19 | 119.5 |
| C6—C7—C8 | 121.38 (14) | C25—C20—C21 | 118.78 (14) |
| C2—C7—C8 | 119.18 (14) | C25—C20—C11 | 119.24 (14) |
| C9—C8—C7 | 121.92 (14) | C21—C20—C11 | 121.95 (14) |
| C9—C8—H8 | 119.0 | C22—C21—C20 | 120.39 (16) |
| C7—C8—H8 | 119.0 | C22—C21—H21 | 119.8 |
| C8—C9—C10 | 120.86 (14) | C20—C21—H21 | 119.8 |
| C8—C9—H9 | 119.6 | C21—C22—C23 | 120.11 (16) |
| C10—C9—H9 | 119.6 | C21—C22—H22 | 119.9 |
| C11—C10—C1 | 117.49 (13) | C23—C22—H22 | 119.9 |
| C11—C10—C9 | 123.43 (13) | C24—C23—C22 | 120.12 (15) |
| C1—C10—C9 | 119.08 (14) | C24—C23—H23 | 119.9 |
| C12—C11—C10 | 118.65 (13) | C22—C23—H23 | 119.9 |
| C12—C11—C20 | 119.37 (14) | C23—C24—C25 | 119.88 (16) |
| C10—C11—C20 | 121.98 (13) | C23—C24—H24 | 120.1 |
| C11—C12—C13 | 120.39 (14) | C25—C24—H24 | 120.1 |
| C11—C12—H12 | 119.8 | C24—C25—C20 | 120.71 (15) |
| C13—C12—H12 | 119.8 | C24—C25—H25 | 119.6 |
| N1—C13—C12 | 121.83 (13) | C20—C25—H25 | 119.6 |
| C13—N1—C1—C10 | 0.0 (2) | C1—N1—C13—C14 | 179.61 (12) |
| C13—N1—C1—C2 | 179.91 (13) | C11—C12—C13—N1 | 0.0 (2) |
| N1—C1—C2—C3 | 0.8 (2) | C11—C12—C13—C14 | −179.01 (13) |
| C10—C1—C2—C3 | −179.29 (14) | N1—C13—C14—C15 | 174.74 (13) |
| N1—C1—C2—C7 | 179.98 (13) | C12—C13—C14—C15 | −6.2 (2) |
| C10—C1—C2—C7 | −0.1 (2) | N1—C13—C14—C19 | −7.5 (2) |
| C7—C2—C3—C4 | −1.5 (2) | C12—C13—C14—C19 | 171.63 (14) |
| C1—C2—C3—C4 | 177.67 (14) | C19—C14—C15—C16 | −0.9 (2) |
| C2—C3—C4—C5 | 1.1 (2) | C13—C14—C15—C16 | 176.90 (14) |
| C3—C4—C5—C6 | 0.2 (2) | C14—C15—C16—C17 | −0.3 (2) |
| C4—C5—C6—C7 | −1.0 (2) | C15—C16—C17—C18 | 1.2 (2) |
| C5—C6—C7—C2 | 0.6 (2) | C15—C16—C17—Cl2 | −178.31 (13) |
| C5—C6—C7—C8 | −179.10 (15) | C16—C17—C18—C19 | −1.0 (2) |
| C3—C2—C7—C6 | 0.7 (2) | Cl2—C17—C18—C19 | 178.52 (11) |
| C1—C2—C7—C6 | −178.51 (13) | C16—C17—C18—Cl1 | −178.77 (12) |
| C3—C2—C7—C8 | −179.61 (14) | Cl2—C17—C18—Cl1 | 0.77 (19) |
| C1—C2—C7—C8 | 1.2 (2) | C17—C18—C19—C14 | −0.2 (2) |
| C6—C7—C8—C9 | 179.24 (15) | Cl1—C18—C19—C14 | 177.60 (12) |
| C2—C7—C8—C9 | −0.4 (2) | C15—C14—C19—C18 | 1.1 (2) |
| C7—C8—C9—C10 | −1.4 (2) | C13—C14—C19—C18 | −176.76 (13) |
| N1—C1—C10—C11 | −1.0 (2) | C12—C11—C20—C25 | 65.91 (19) |
| C2—C1—C10—C11 | 179.08 (13) | C10—C11—C20—C25 | −113.34 (17) |
| N1—C1—C10—C9 | 178.24 (13) | C12—C11—C20—C21 | −112.36 (17) |
| C2—C1—C10—C9 | −1.7 (2) | C10—C11—C20—C21 | 68.4 (2) |
| C8—C9—C10—C11 | −178.35 (14) | C25—C20—C21—C22 | 0.2 (2) |
| C8—C9—C10—C1 | 2.5 (2) | C11—C20—C21—C22 | 178.44 (15) |
| C1—C10—C11—C12 | 1.5 (2) | C20—C21—C22—C23 | 0.1 (3) |
| C9—C10—C11—C12 | −177.71 (14) | C21—C22—C23—C24 | 0.3 (3) |
| C1—C10—C11—C20 | −179.25 (13) | C22—C23—C24—C25 | −0.9 (3) |
| C9—C10—C11—C20 | 1.5 (2) | C23—C24—C25—C20 | 1.2 (2) |
| C10—C11—C12—C13 | −1.1 (2) | C21—C20—C25—C24 | −0.8 (2) |
| C20—C11—C12—C13 | 179.67 (13) | C11—C20—C25—C24 | −179.11 (14) |
| C1—N1—C13—C12 | 0.5 (2) |
Hydrogen-bond geometry (Å, °)
| Cg1, Cg2 and Cg3 are the centroids of the C20–C25, C14–C19 and N1/C1/C10–C13 rings, respectively. |
| D—H···A | D—H | H···A | D···A | D—H···A |
| C6—H6···Cg1i | 0.95 | 2.98 | 3.8577 (19) | 154. |
| C22—H22···Cg2ii | 0.95 | 2.94 | 3.8204 (19) | 156. |
| C25—H25···Cg3iii | 0.95 | 2.63 | 3.4738 (17) | 148. |
| C19—H19···N1 | 0.95 | 2.42 | 2.765 (2) | 101. |
Symmetry codes: (i) x, −y+3/2, z−1/2; (ii) x−1, y, z; (iii) −x, −y+2, −z+2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HG5101).
References
- Cho, Y. J., Kwon, H. J., Kim, B. O., Kim, S. M. & Yoon, S. S. (2010). Eur. Patent No. 2182002.
- Liu, J., Leung, C.-H., Chow, A. L.-F., Sun, R. W.-Y., Yan, S.-C. & Che, C.-M. (2011). Chem. Commun. 47, 719–721. [DOI] [PubMed]
- Pantoom, S., Vetter, I. R., Prinz, H. & Suginta, W. (2011). J. Biol. Chem. 286, 24312–24323. [DOI] [PMC free article] [PubMed]
- Rigaku/MSC (2002). CrystalClear Rigaku/MSC Inc., The Woodlands, Texas, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Zhang, Y., Li, P. & Wang, L. (2010). J. Heterocycl. Chem. 48, 153–157.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536811040049/hg5101sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811040049/hg5101Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811040049/hg5101Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




