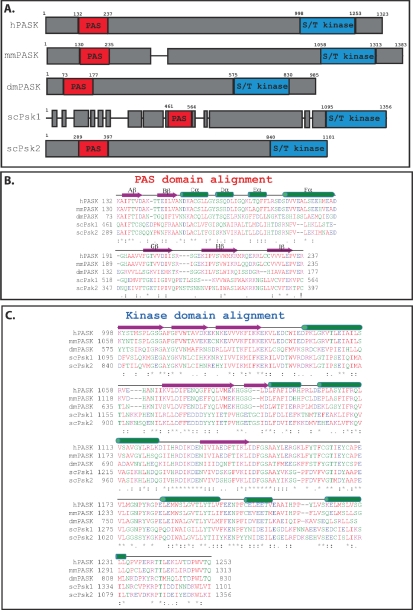
Figure 2.
Alignment of the PAS and kinase domains from selected PAS kinase orthologs. (A) Schematic of PAS kinase homologs. Regions of similarity are boxed in grey, with non-homologous regions indicated by gaps between the grey boxes. Alignment of the PAS (B) and kinase (C) domains from selected PAS kinase orthologs. Secondary structure elements (α-helical and β-sheet) were derived from the published NMR structure of the PAS domain [27] as well as amino acid alignment of the kinase domain with the published structure of microtubule-associate protein/microtubule affinity regulating kinase 2 (MARK2) [33], and are shown above the amino acids in green and purple respectively. Amino acid sequences were aligned using the Clustal W program [34]. The degree of amino acid conservation is indicated both by color and “*” (identical residues in all sequences), “:” (highly conserved amino acids), and “.” (weakly conserved amino acids). Red indicates a small hydrophobic or aromatic amino acid (-Y), blue indicates acidic, magenta is basic, and green is hydroxyl plus amine plus basic (-Q), all other amino acids are grey.