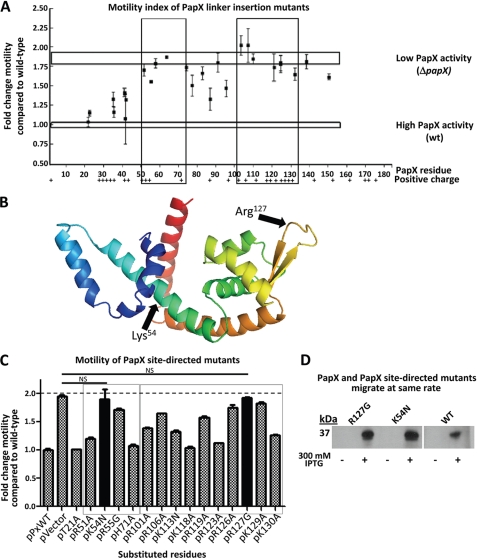
FIGURE 4.
Motility index of 15-bp insertional mutants of papX. A, 30 in-frame 15-bp insertions were made in papX in pPxWT. The 30 constructs, pPxWT (WT papX), and pVector (empty vector) were electroporated into CFT073 ΔpapX. Motility was assessed in soft agar and compared relative to CFT073 ΔpapX electroporated with pPxWT. Vertical boxes indicate regions of loss of function. Top and bottom horizontal boxes represent motility of CFT073 ΔpapX electroporated with pVector or with pPxWT, respectively. The presence of positively charged residues is indicated along the bottom. B, I-TASSER software was used to generate a predicted structural model based on the amino acid sequence of PapX. Arrows indicate putative DNA binding (Arg127) and dimerization (Lys54) regions of the protein. C, site-directed mutants of papX were made in pPxWT and transformed into a CFT073 ΔpapX background. Soft agar motility assays were performed on the resulting constructs, performed in triplicate using biological triplicates. Boxes indicate regions sensitive to mutation determined in A by linker-insertional mutagenesis. Solid bars are not significantly different from vector control (dashed line). D, Western blot showing pPxWT (WT PapX), pR127G (PapX R127G mutant), and pK54N (PapX K54N mutant) transformed into E. coli strain K12 either uninduced or induced with 300 mm IPTG. Bands migrate with the same electrophoretic mobility for mutants and wild-type PapX by seminative SDS-PAGE (12% acrylamide). Error bars, S.E.