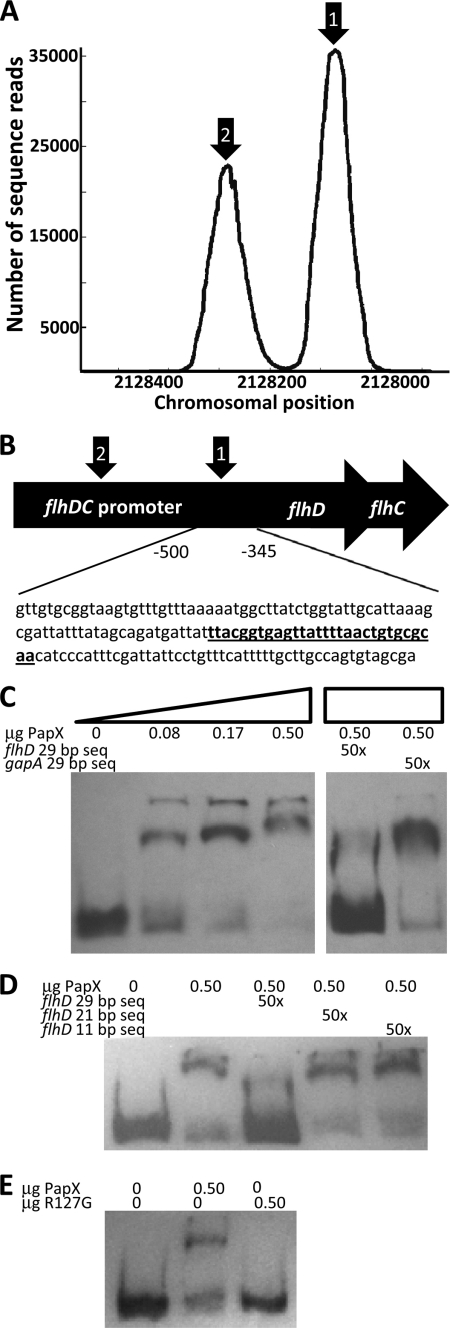
FIGURE 8.
PapX binds to the flhDC promoter. A, dot plot of chromosomal frequencies by SELEX revealed two significant peaks within the flhDC promoter (arrows 1 and 2). The peak under arrow 1 includes the predicted PapX binding site. B, schematic of the flhDC promoter, indicating the region −346 to −501 bases upstream of the flhD translational start used in subsequent gel shifts. A perfectly identical (29 of 29) predicted PapX binding site is indicated in underlined boldface type and is centered 410 bp upstream of the flhD translational start site. C, increasing amounts of PapX protein were added to 3.2 ng of DIG-UTP-end-labeled 154-base pair fragments derived from the flhDC promoter, the sequence shown in B. Unlabeled competitor DNA at 50-fold excess by mass was added to the protein-DNA binding reaction for each of the last two lanes. flhD 29 bp seq, sequence indicated in underlined boldface type in B. gapA 29 bp seq, sequence ACTCCACTCACGGCCGTTTCGACGGTACCG from gapA. D, the competition assay performed in C was repeated using only the 0.50-μg amount of PapX-His6 protein. Additional sequences derived from the 29-bp competitor (TTACGGTGAGTTATTTTAAC, indicated by flhD 21 bp seq, and GTTATTTTAAC, indicated by flhD 11 bp seq), determined by sequence analysis in Fig. 7, D and E, were also used in 50-fold excess by mass. E, gel shifts were repeated with the same 154-bp fragment as in C and with 0.50-μg amounts of wild-type PapX-His6 protein and PapX-His6 containing the R127G mutation.