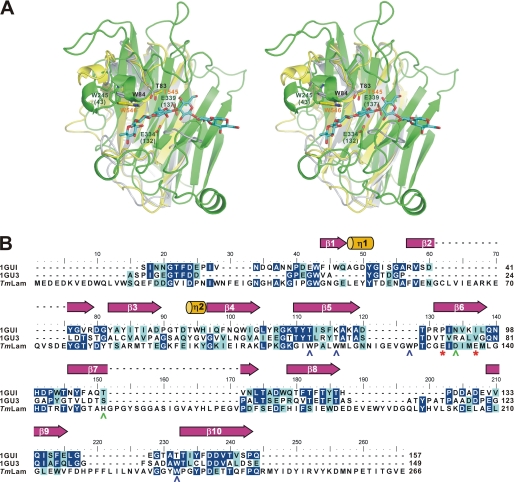
FIGURE 4.
Comparison of the carbohydrate-binding modules and the catalytic domain of TmLam. A, TmLamCD (chain C of 3AZZ, in green), TmLamCBM2 (1GUI, in yellow), and the modeled TmLamCBM1 (in gray) were superposed using PDBeFold server. The modeled TmLamCD-laminarihexaose molecule was then aligned with the superposed TmLamCD using PyMOL. The laminarihexaose is shown as cyan sticks. Amino acid residues are labeled in accordance with the overall TmLam protein numbering and with TmLamCD in parentheses. Residues belonging to TmLamCBM1 are labeled in black, those for TmLamCD are in dark green, and those for TmLamCBM2 are in orange. B, ESPript produced figure using the aligned sequence by ClustalW. 1GU3, CBM4–1 from Cellulomonas fimi; TmLam, the catalytic domain of TmLam. The catalytic Glu residues are denoted as red asterisks below the TmLam sequence and shown as green sticks in A. The secondary structure of the 1GUI is assigned above. Amino acids of TmLamCD with polar side chains around the active site are labeled in green, whereas aromatic amino acids are in blue.