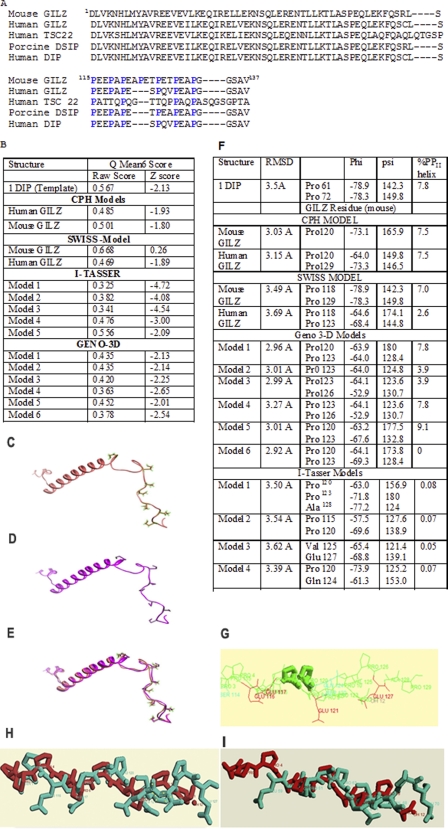
FIGURE 1.
A, sequence alignment of human GILZ (BAB18680.1), mouse GILZ (AAD01789.1), human TSC22 (CAA10951.1), porcine DSIP (AAB28177.1), and human DIP by CLUSTAL W (46). The conserved proline residues are highlighted. Homology modeling of mouse GILZ and human GILZ were generated based on sequence and structure similarity with the porcine δ sleep-inducing peptide (PDB 1DIP). B, validity of the models from CPH Models, Geno3D, Swiss Model, I-Tasser, and Geno3D were assessed with the QMEAN 6 program. The QMEAN 6 score is a composite score consisting of a linear combination of 6 terms (C-β interaction energy, all atom pairwise energy, salvation energy, torsion angle energy, secondary structure agreement, and solvent accessibility agreement). The pseudo-energies of the contributing terms (raw scores) are given together with their Z-scores with respect to scores obtained for high-resolution experimental structures of similar size solved by x-ray crystallography. C and D show the structure of PDB code 1DIP and a representative model of the mouse GILZ generated by Swiss Model. E, comparison by overlap yields a root mean square deviation of less than 0.1 Å between the mouse GILZ model and PDB code 1DIP suggesting excellent structural symmetry. F shows the values of the backbone dihedral angles depicting the secondary structure of the GILZ-COOH as assigned by PROSS. The torsion angles φ and π of Pro120 and Pro123 are consistent with the PPII helical conformation in the predicted GILZ models. The root mean square deviation of the superimposition of the GILZ models against the x-ray crystal structure of the synthetic PPII helix as determined by CHEMERA is also given. G–I show representative superimposed structures of synthetic PPII in gray and the (G) CPH mouse-GILZ model, (H) Swiss Model mouse GILZ model, and (I) Geno-3D mouse GILZ model in blue. Pro120 and Pro123 are highlighted.