FIGURE 6.
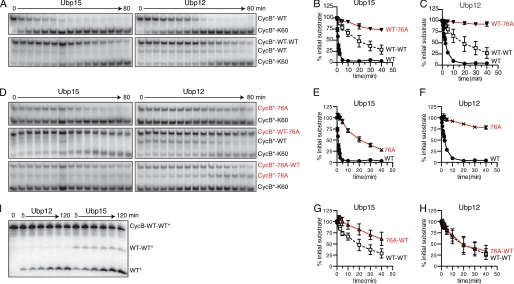
Ubp15 and Ubp12 show distinct base- and tip-directed activities. A, Ubp15 and Ubp12 deubiquitination of mono- (top panels) or diubiquitinated CycB (bottom panels). Time course of 125I-labeled CycB-Lys60 deubiquitination by 0.4 nm Ubp15 (left) or 0.5 nm Ubp12 (right). K48R ubiquitin was used for monoubiquitination and as the distal ubiquitin of diubiquitin chains. APC/C reactions with Ubc4 attached the first ubiquitin to CycB; Ubc1 was substituted for Ubc4 to attach a second ubiquitin. APC/C reactions were terminated with heating. B and C, quantitation of data from A and D. Levels of each ubiquitin conjugate are normalized to their initial values (in arbitrary units) at time zero. Error bars denote S.E. from two to three replicates. D, G76A ubiquitin conjugates reveal base versus tip selectivity. Shown is the time course as described in A, substituting ubiquitin bearing G76A where indicated. E–H, quantitation of data from A and D, as described in B. I, time course of CycB diubiquitin hydrolysis in which the distal ubiquitin, rather than CycB, is radiolabeled. K48R ubiquitin bearing a PKA phosphorylation site was ligated to wild-type ubiquitin with Cdc34 and purified by ion exchange chromatography. After phosphorylation, this diubiquitin was ligated to unlabeled CycB-Lys60 in an APC/C reaction with Ubc4. After gel purification, 50 nm CycB diubiquitin was incubated with 200 nm Ubp12 (left) or Ubp15 (right). See supplemental Table S1 for initial rates.