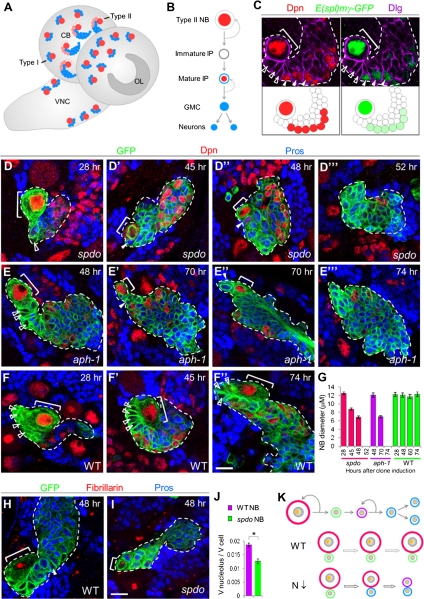
Figure 1.
N-dependent cell growth is required for type II NB maintenance. (A) A schematic drawing of Drosophila late larval CNS showing type I and type II NB lineages within the central brain area. (CB) Central brain; (OL) optical lobe; (VNC) ventral nerve cord. (B) A diagram of a type II NB lineage. Distinct cell types within the hierarchy can be identified by using combinations of cell fate markers: type II NB: Dpn (Deadpan, red)+, Pros (Prospero, blue)−; immature IPs: Dpn−, Pros−; mature IPs: Dpn+, cytoplasmic Pros; GMC or neurons: Dpn−, nuclear Pros. (C) N reporter E(spl)mγ-GFP (green) expression in type II NB lineage (mature IPs, closed arrowheads) showing differential N activity in different cell types. Dlg staining (purple) outlines the cell cortex. (White closed arrowheads) Mature IPs. From this panel on, NBs are marked with brackets and immature IPs are marked with white open arrowheads. (D–G) Clonal analysis of type II NBs of spdo (D) or aph-1 (E) mutants at various times ACI. Newly born daughter cells are marked with closed arrowheads. (F) Wild-type NBs served as control. (H,I) Analysis of the nucleoli (red; anti-fibrillarin) of the NBs and their daughter cells within spdo mutant (I) or wild-type (H) clones. (J) Quantification of nucleolar/cellular volume ratio of wild-type or spdo mutant NBs. (*) P < 0.002 versus control in Student's t-test; n = 6–8. (K) A working model depicting N regulation of NB fate through control of their cellular and nucleolar sizes. (Orange dot) Nucleolus; (red) NB; (green) immature IP; (purple) mature IP; (blue) GMCs or neurons. Bar, 10 μm.