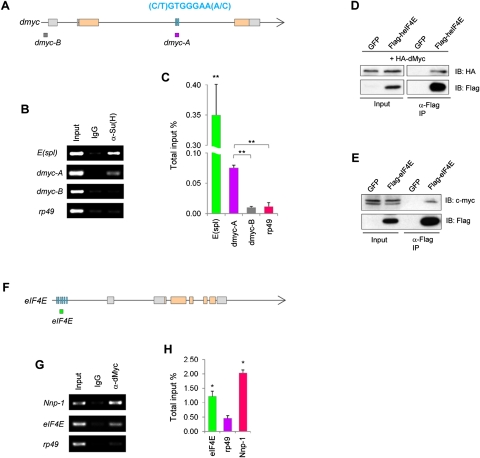
Figure 5.
Biochemical characterization of the N-dMyc–eIF4E molecular circuitry. (A–C) ChIP with the anti-Su(H) antibody or a control IgG on wild-type third instar larval brain chromatin. (A) Schematic representation of the dmyc locus. (Orange rectangles) Coding regions; (gray rectangles) noncoding regions; (lines) introns; (blue bars) two putative Su(H)-binding sites matching the consensus sequence (C/T)GTGGGAA(A/C). Enrichment of Su(H) at the dmyc-A but not dmyc-B amplicon is determined by both standard PCR (B) and real-time quantitative PCR (C). A region in E(spl) containing Su(H)-binding sites (Bailey and Posakony 1995) and a region in the rp49 promoter without such sites serve as positive and negative controls, respectively. (**) P < 0.0001. (D,E) Coimmunoprecipitation between transfected human eIF4E and Drosophila dMyc (D) or human eIF4E and endogenous c-Myc (E) in HEK293T nuclear extracts. GFP serves as a negative control. (IP) Immunoprecipitation; (IB) immunoblotting. Input represents 4% of total. (F–H) ChIP with anti-dMyc antibody or a control IgG on wild-type third instar larval brain chromatin. (F) Schematic representation of the eIF4E locus. Blue bars represent a cluster of adjacent noncanonical E boxes. Enrichment of dMyc at the eIF4E locus is shown by standard PCR (G) and real-time quantitative PCR (H). A region in Nnp-1 harboring canonical E boxes CACGTG and a region within the rp49 promoter serve as positive and negative controls, respectively. (*) P < 0.01. Error bars indicate standard deviation of three independent experiments.