Figure 6.
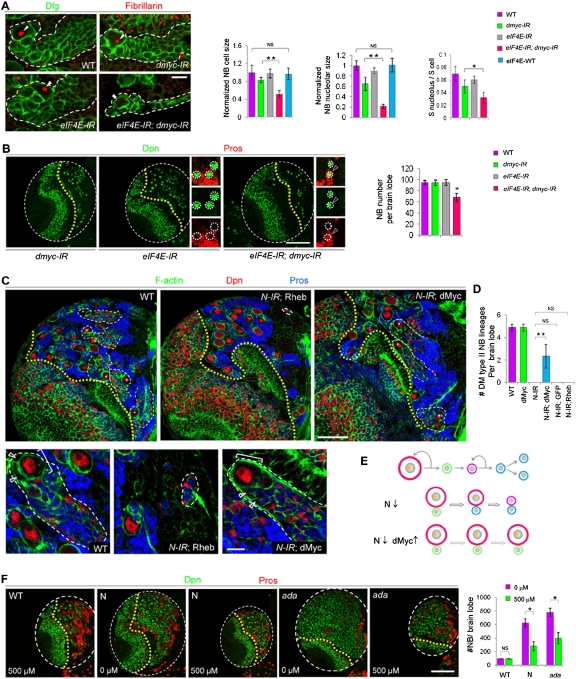
The N-dMyc–eIF4E module operates in normal or ectopic NBs. (A) dMyc or eIF4E knockdown alone has a mild or no effect on NB cell growth, respectively, as assayed by their cell size, nucleolar size, or ratio of nucleolar/cell size, whereas knocking down both leads to a drastic reduction in NB cell growth. Overexpression of a wild-type form of the eIF4E transgene shows no discernible effect on NB cellular or nucleolar sizes. NB nucleoli are indicated by arrowheads. (Green) Dlg; (red) fibrillarin. (**) P < 0.0001; (*) P < 0.05; n = 30–40. (B) Knocking down either dMyc or eIF4E alone has no noticeable effect on type II NB maintenance, but knocking down both leads to premature differentiation and NB loss. (Green) Dpn; (red) Pros. (*) P < 0.001; n = 15–20. Magnified images are shown on the right, with individual NBs encircled with white dotted lines. (Arrowheads) Pros+ Dpn+ cell. (C) Type II NB lineages in wild-type, 1407>N RNAi; Rheb, or1407>N-IR; dMyc backgrounds are delineated by white dashed lines, and the NB in each lineage is indicated by a star. The yellow dotted line indicates the boundary between the optical lobe (left) and the central brain (right) region. Amplified images of a representative type II NB lineage in each genotype are shown at the bottom. Larvae were processed at 96 h ALH. (D) Quantification of type II NB number in wild-type, 1407>dMyc, 1407>N-IR, 1407>N-IR; dMyc, 1407>N-IR; GFP, or 1407>N-IR; Rheb brain lobes. (**) P < 0.0005; n = 15–20. Only dorsal–medial (DM) type II NBs generating surface layer progeny were counted. (E) A working model of dMyc overexpression preventing the stem cell fate loss in N signaling-defective NBs by boosting cell growth. (Orange dot) Nucleolus; (red) NB; (green) immature IP; (purple) mature IP; (blue) GMCs or neurons. (F) Effects of eIF4E inhibitor treatment on wild-type NB maintenance and ectopic NB formation induced by N overactivation or Ada inactivation. (*) P < 0.0001; n = 15–20. Bars: A,C, bottom panel, 10 μm; C, top panel, 50 μm; B,F, 100 μm.