Summary
Tell mediated immunity requires T cell receptor (TCR) cross-reactivity, the mechanisms behind which remain incompletely elucidated. The αβ TCR A6 recognizes both the Tax (LLFGYPVYV) and Tel1p (MLWGYLQYV) peptides presented by the human class I MHC molecule HLA-A2. Here we found that although the two ligands are ideal structural mimics, they form substantially different interfaces with A6, with conformational differences in the peptide, the TCR, and unexpectedly, the MHC molecule. The differences between the Tax and Tel1p ternary complexes could not be predicted from the free peptide-MHC structures and are inconsistent with a traditional induced-fit mechanism. Instead, the differences were attributable to peptide and MHC molecular motion present in Tel1p-HLA-A2 but absent in Tax-HLA-A2. Differential “tuning” of the dynamic properties of HLA-A2 by the Tax and Tel1p peptides thus facilitates cross-recognition and impacts how structural diversity can be presented to and accommodated by receptors of the immune system.
Introduction
Recognition of an antigenic peptide-MHC complex by an αβ T cell receptor (TCR) initiates an intracellular signaling cascade leading to a T cell response. Although antigen specificity is considered a hallmark of cellular immunity, the TCR is cross-reactive, binding and responding to multiple peptide-MHC (pMHC) ligands. Molecular mimicry, in which cross-reactive ligands share key structural and chemical features(Kohm et al., 2003), is a mechanism commonly used to explain TCR cross-reactivity. In principle, molecular mimicry allows a single TCR to engage cross-reactive ligands similarly. Yet molecular mimicry cannot easily explain cross-reactive recognition of ligands with little or no structural or chemical homology. A recently identified mechanism facilitating such cross-recognition is the global repositioning of a receptor on the surface of different ligands, allowing the formation of stable interfaces despite imperfect homology (Colf et al., 2007). Another mechanism to explain cross-reactive recognition of dissimilar ligands involves conformational changes and flexibility in the TCR complementary determining region (CDR) loops, which can permit the TCR to adapt structurally to different pMHC ligands (Gagnon et al., 2006; Mazza et al., 2007).
Yet in addition to the TCR, conformational shifts in both peptide and MHC have been shown to occur upon TCR binding (Garboczi et al., 1996; Ishizuka et al., 2008; Rudolph et al., 2001; Tynan et al., 2007). Excluding the extensively bulged EPLP peptide(Tynan et al., 2007), these shifts are frequently small and usually interpreted as arising from “induced-fit” type conformational adjustments. However, structurally observed conformational shifts occurring upon protein binding can result from inherent molecular flexibility (James et al., 2003; Lange et al., 2008; Tobi and Bahar, 2005), and recent studies have shown how subtle differences in class I MHC molecules can lead to different peptide dynamics in MHC binding grooves (Arch bold et al., 2009; Fabian et al., 2008; Pullman et al., 2004). Given the growing realization that even small amounts of flexibility can profoundly influence molecular recognition, signaling, and ultimately biological function (Smock and Gierasch, 2009), a closer examination of the role of pMHC flexibility and its potential to influence immune recognition is warranted.
The human αβ TCR A6 recognizes the HTLV-1 Tax peptide (LLFGYPVYV) presented by the class I MHC molecule HLA-A*0201 (HLA-A2). In addition to the Tax peptide, A6 also recognizes the Saccharomyces cerevisiae peptide Tel1p (MLWGYLQYV). Tel1p was identified as a ligand for A6 after screening databases for Tax-like peptides which matched the A6 recognition motif (Hausmann et al., 1999). Cross-reactivity between Tax and Tel1p is not unexpected given the similarities in the peptides.
However, differences in the fine specificity of A6 towards Tax and Tel1p (Laugel et al., 2005) prompted us to examine the recognition of the Tel1p ligand in greater detail. Surprisingly, we found that although the free Tel1p-HLA-A2 complex is an ideal structural mimic of the Tax ligand, the interface A6 forms with Tel1p-HLA-A2 differs substantially from the interface it forms with Tax-HLA-A2, with differences in the peptide, the TCR CDR3 β loop, and unusually, the HLA-A2 α2 helix. The conformational differences between free and bound Tel1p-HLA-A2 and the changes in the A6 CDR3β loop changes are interdependent, indicating that TCR recognition of the Tel1p ligand requires the mutual adaptation of both the TCR and the pMHC binding surfaces. Notably, the conformational changes are inconsistent with a purely induced-fit binding mechanism. Rather, in line with emerging data on the influence of protein dynamics in molecular recognition, molecular motion present in unligated Tel1p-HLA-A2 but absent in unligated Tax-HLA-A2 was identified as the chief contributor to the differences between A6 recognition of Tel1p and Tax, with the different degrees of flexibility permitting the formation of different but equally complementary TCR-pMHC interfaces. These findings shed light on how structural diversity can be presented to and accommodated by receptors of the immune system, and reveal that antigen-dependent “tuning” of molecular motion distributed throughout the TCR binding surface of the pMHC can contribute to TCR recognition and facilitate cross-reactivity.
Results
The Tel1p-HLA-A2 and Tax-HLA-A2 complexes are ideal structural mimics
We first determined the structure of the Tel1p-HLA-A2 complex at 1.9 Å resolution(Fig. 1). The complex crystallized isomorphously with the 1.8 Å Tax-HLA-A2 complex (Khan et al., 2000). Data collection and refinement statistics are in Table 1; electron density images are in Supplemental Figure S1. The HLA-A2 heavy chain in Tel1p-HLA-A2 adopts the usual class I MHC architecture, and the peptide-binding domain superimposes onto that of the Tax-HLA-A2 structure with a backbone RMSD of 0.3 Å (Fig 1). The N-and C-terminal regions of the Tel1p and Tax peptides adopt similar conformations in the peptide binding groove, and the side-chains of Tyr5 and Tyr8 are super imposable (a water molecule was modeled adjacent to Tyr5; we considered whether this could reflect an alternate conformation for the Tyr5 side chain, yet the side chain and backbone density in both traditional and omit maps as well as sterics indicated the side chain position shown in Fig. 1 was correct).
Figure 1.
The unligated Tel1p and Tax complexes with HLA-A2 are sequence and structural mimics. A) Sequences of the Tax and Tel1p peptides. B) Structure of the unligated Tel1p-HLA-A2 peptide-binding domain superimposed on the Tax-HLA-A2 peptide binding domain. Superimposition is through the backbones of the peptide-binding domains. The color code is given at the bottom of the figure and maintained in panels C and D. C) Superimposition of the peptides from the free Tel1p-HLA-A2 structure and the Tax peptide from the free Tax-HLA-A2 structure. D) Superimposition of the peptides from the free Tel1p-HLA-A2 structure and the Tax peptide from the A6–Tax-HLA-A2 structure. Superimposition for all panels is by backbone atoms.
Table 1.
X-ray data and refinement statistics
| Protein complex | Tel1p-HLA-A2 | A6–Tel1p-HLA-A2 | Tel1p-HLA-A2 (A150P) | Tax-HLA-A2 (A150P) |
|---|---|---|---|---|
| PDB entry | 3H7B | 3H9S | 3H9H | 3IXA |
| Radiation source | APS 19BM | APS 19BM | APS 31ID | APS 31ID |
| Space group | P1 | C2 | P21 | P21 |
| a [Å] | 50.32 | 223.11 | 63.16 | 60.89 |
| b [Å] | 63.06 | 48.31 | 88.89 | 85.29 |
| c [Å] | 74.88 | 92.48 | 79.41 | 82.48 |
| α [°] | 81.95 | 90 | 90 | 90 |
| b [°] | 75.95 | 90.67 | 89.97 | 89.84 |
| γ [°] | 77.88 | 90 | 90 | 90 |
| Molecules/a.u. | 2 | 1 | 2 | 2 |
| Resolution [Å] | 20 – 1.9 | 20 – 2.7 | 20 – 2.0 | 20 – 2.1 |
| Total number of reflections | 67439 | 27920 | 56073 | 48760 |
| Mosaicity [°] | 0.53 | 0.89 | 0.26 | 0.96 |
| Completeness a [%] | 96.8 (94.4) | 99.4 (98.2) | 97.3 (86.3) | 97.1(76.3) |
| I/σ | 13.5 (1.98) | 17.7 (2.0) | 18.0 (2.6) | 19.4(1.73) |
| Rmerge [%] | 6.6 (35.6) | 8.8 (45.3) | 6.9 (31.6) | 6.4(43.1) |
| Average redundancy | 1.9(1.9) | 3.5 (3.1) | 3.6 (2.9) | 3.5(2.2) |
| Rwork [%] (no. reflections) | 19.2 (64026) | 21.7 (25991) | 17.1 (53214) | 18.8(46274) |
| Rfree [%] (no. reflections) | 24.2 (3396) | 28.9 (1387) | 22.6 (2841) | 23.0 (2464) |
| Average B factor (all atoms) [Å2] | 26.2 | 62.8 | 36.5 | 23.2 |
| Ramachandran plot | ||||
| Most favored [%] | 91.6 | 86.9 | 92.8 | 92.3 |
| Allowed [%] | 8 | 12.4 | 7 | 7.4 |
| Generously allowed [%] | 0.4 | 0.6 | 0.1 | 0.3 |
| RMS deviations from ideality | ||||
| Bonds [Å] | 0.016 | 0.013 | 0.016 | 0.017 |
| Angles [°] | 1.717 | 1.675 | 1.632 | 1.927 |
| Coordinate error b [Å] | 0.13 | 0.33 | 0.12 | 0.14 |
Numbers in parenthesis refer to the highest resolution shell
Mean estimate based on maximum likelihood methods
The backbones of the Tax and Tel1p peptides diverge at positions 6 and 7, but when the Tel1p peptide is compared to the Tax peptide from the A6–Tax-HLA-A2 complex, in which the peptide backbone is “squished” at positions 6 and 7(Garboczi et al., 1996), the backbones are in full alignment (the RMSD for superimposition of all atoms of Tel1p onto all atoms of unligated Tax is 0.8 Å; using ligated Tax the value is 0.4 Å) (Fig 1B–D). Tel1p-HLA-A2 is thus an ideal structural mimic of Tax-HLA-A2.
Recognition of Tel1p-HLA-A2 proceeds via conformational changes in the peptide, TCR, and MHC
We next determined the structure of the A6–Tel1p-HLA-A2 complex (Fig. 2). The crystals diffracted to 2.7 Å resolution and were of the same form as those of A6 bound to native Tax and six Tax variants (Ding et al., 1999; Gagnon et al., 2006; Garboczi et al., 1996; Piepen brink et al., 2009). Data collection and refinement statistics are in Table 1; electron density images are in Supplemental Figure S2. The binding mode of A6 on Tel1p-HLA-A2, including the docking angle and tilt, is the same with Tel1p as with Tax (Fig. 2A). However, upon examining the TCR-pMHC interface, differences were seen in the conformation of the peptide, the TCR, and most unusually, HLA-A2 (Fig. 2B).
Figure 2.
Recognition of Tel1p-HLA-A2 by the A6 TCR proceeds with changes in the peptide, the TCR, and the HLA-A2 α2 helix. The color code for all panels is given in the lower right of the figure. A) Overview of the A6–Tel1p-HLA-A2 and A6–Tax-HLA-A2 complexes. B) Cross-eyed stereo view highlighting the three major changes in the interface: peptide, TCR CDR3β, and HLA-A2 α2 helix. C) Close-up view of the changes that occur in the Tel1p peptide upon TCR binding. The rotations of the Tyr5 and Gln7 side-chains are indicated, as is the distance moved and the compensating rotations in the Tyr5 ψ and the Leu6 φ bonds. The clash that would occur between the Tyr5 side-chain and the peptide backbone if the backbone did not rotate is indicated in red. D) Close-up of the difference in the HLA-A2 α2 helix between the Tel1p and Tax complexes with A6. The differential position of the Ala150 carbon in the two complexes is indicated, and the shifting region of the helix is in red. E) Close-up of the difference in the A6 CDR3β loop between the Tel1p and Tax complexes with A6. The differential position of the carbon of Gly101β in the two complexes is indicated. F) The various conformational changes are mutually dependent. Steric clashes occur if the Tel1p peptide from the Tel1p ternary complex is fit into the peptide-binding domain from the unligated Tel1p-HLA-A2 complex or into the A6–Tax-HLA-A2 interface. Clashes, defined as interatomic distances less than the sum of the corresponding van der Waals radii, are shown as dashed red lines. Superimposition for all panels is through backbone atoms of the variable and peptide-binding domains.
A defining feature of the A6 TCR bound to the Tax peptide is the “pocket” formed between the CDR3α and CDR3β loops(Garboczi et al., 1996; Hausmann et al., 1999). In recognition of the Tax peptide by A6, Tyr5 maintains the conformation seen in the unbound pMHC and occupies this pocket. In the structure with Tel1p, however, Tyr5 has moved away from its unbound conformation and now points towards the HLA-A2 α2 helix (Fig. 2C). The movement occurs via a 100° rotation around χ1 and results in a 9.6 Å displacement of the tyrosine hydroxyl (weak electron density was observed near the position the Tyr5 ring occupies in the Tax structure; this was modeled as a water as both traditional and omit maps indicated stronger and more complete side chain density for the position shown in Fig. 2C).
Rotation of the Tel1p Tyr5 χ1 angle is permitted by an 88° rotation in the Tyr5 ψ bond (Fig. 2C), which points the Tyr5 carbonyl oxygen towards the base of the peptide binding groove. Without this rotation, the movement in the side-chain would be prevented by an overlap with the carbonyl oxygen. Following rotation of the Tyr5 ψ bond, the path of the peptide is retained in an extended conformation by an 89° counter-rotation of the Leu 6 φ bond. Along with the rotation of Tyr5, the Tel1p Gln7 side-chain rotates by 62° around χ1. This shifts the Gln7 side-chain away from Tyr5 by 2.4 Å, likely to minimize repulsion between the oxygen atoms of the side-chains.
Along with the changes in the peptide, a surprising structural reorganization in HLA-A2 occurs upon A6 recognition of Tel1p-HLA-A2 (Fig. 2D). The change occurs in and around the small loop comprised of Ala150-Val152 that normally links the short and long helical elements of the α2 helix. The change is best summarized as a conformational “switch” that shortens the short arm of the helix and extends the long arm of the helix, such that Ala150, which is normally at the C-terminus of the short helix, now forms the N-terminus of the long helix. Measured by the position of its α carbon, Ala150 moves by 6.9 Å. To accommodate its new N-terminus, the long helix is displaced away from the peptide and does not return to its normal position until Gln155, one-third down the length of the long segment of the α2 helix. The short helical segment, rather than ending at Ala150, now ends three residues earlier at Trp147. The switch in the α2 helix is necessary to accommodate the changes in the Tel1p peptide: without the switch, overlap would occur between atoms within the peptide and the α2 helix (Fig. 2F).
Along with the changes in the peptide and the MHC, the CDR3β loop of the A6 TCR in the A6–Tel1p-HLA-A2 complex is shifted up and away from its position in the A6–Tax-HLA-A2 complex. The shift is maximal at the Gly101β α carbon, resulting in a 4.5 Å displacement for the apex of CDR3β (Fig. 2E). As with the change in the MHC α 2 helix, the shift in CDR3β is necessary to accommodate the changes in the Tel1p peptide, in this case to avoid steric clashes between atoms of the loop and Tyr5 (Fig. 2F). The shift in CDR3β is similar to, albeit slightly smaller than, the CDR3β shift seen upon A6 recognition of the bulky Tax-5K-IBA peptide (Gagnon et al., 2006)(Fig. 3A), resulting in a large distortion of the central pocket between the CDR3α and CDR3β loops (Fig. 3B). The other five CDR loops remain in conformations similar to those seen in the Tax structure, with maximum displacements of 1 Å for CDR3α and CDR2α.
Figure 3.
Recognition of Tel1p by A6 proceeds with changes in the CDR3β loop, but other loops are largely unaltered. A) Cross-eyed stereo view of the CDR loop positions in the complexes of A6 with the Tel1p, Tax, and Tax-5K-IBA peptides (color code is under the panel). The view is through the peptide-binding domain of HLA-A2, with the Tel1p peptide shown in magenta for reference. Compared to recognition of the Tax peptide, the large shift in CDR3β upon recognition of Tel1p is evident, and it is similar but slightly smaller than the shift seen in recognition of Tax-5K-IBA (Gagnon et al., 2006). B) Comparison of the TCR central pocket in the Tel1p (left) and Tax (right) ternary complexes. The view is as in panel A, with the surface of the TCR shown. CDR3β is blue and CDR3α is yellow. Superimposition for both panels is by the backbones of the variable domains.
The interfacial contacts in the A6–Tel1p-HLA-A2 structure are significantly altered compared to the structure with Tax. Relative to the Tax structure, with Tel1p there is a net loss of seven TCR-pMHC hydrogen bonds, including three between CDR3β and the HLA-A2 α2 helix. This includes a hydrogen bond to Gln155 of HLA-A2, which has been proposed to serve as a minimal TCR “anchor” point on class I MHC molecules(Clements et al., 2006). The contacts to the α 2 helix linker region are substantially altered, and several amino acids that figure prominently in the Tax interface participate in the Tel1p interface minimally or not at all (a detailed comparison of the two interfaces is given in Supplemental Figure S3). Notably, however, the shape complementarity between the two interfaces is unchanged: both the A6–Tel1p-HLA-A2 and A6–Tax-HLA-A2 interfaces have shape complementarity statistics of 0.63 (Lawrence and Colman, 1993).
The reasons for the conformational differences between the interfaces A6 forms with Tel1p-HLA-A2 and Tax-HLA-A2 are not immediately apparent from examining the structures. This is clear when the unligated structure of Tel1p-HLA-A2 is superimposed onto the structure of Tax-HLA-A2 in the A6–Tax-HLA-A2 ternary complex. As shown in Supplemental Figure S4, the steric clashes that occur in this superimposition are not extensive and could be eliminated by more subtle shifts of the side chains of Gln155 of HLA-A2 and Gln7 and Tyr8 of the Tel1p peptide. There are no crystallographic contacts to the centers of the peptides or the region of the HLA-A2 α 2 helix spanning amino acids 146–156 in any of the structures examined.
Altogether, the structural data indicate that A6 forms distinctly different yet equally complementary interfaces with the Tel1p-HLA-A2 and Tax-HLA-A2 ligands. That this occurs despite a high degree of structural and sequence homology between the unligated pMHC complexes suggests the influence of a dynamic mechanism not discernable from structure alone.
Mutagenesis confirms the switch in HLA-A2 and suggests coupling between Tel1p and HLA-A2 flexibility
Although small rigid body displacements of the class I MHC α1 and/or α2 helices have been previously observed to occur upon TCR binding (Ishizuka et al., 2008; Rudolph et al., 2001), the structural remodeling observed upon A6 recognition of Tel1p-HLA-A2 was unusual and unanticipated (Supplemental Fig. S5). We thus sought to independently confirm the conformational switch in the HLA-A2 α2 helix that occurs upon A6 recognition of Tel1p-HLA-A2.
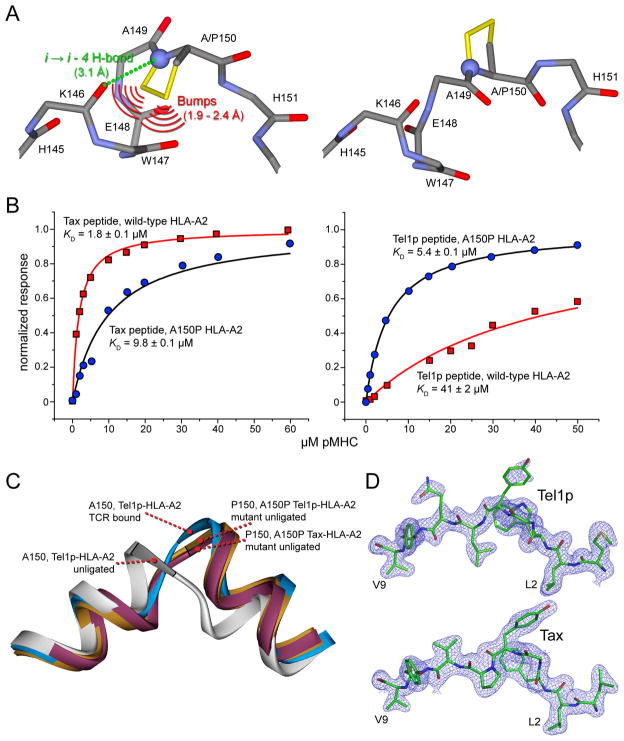
To do this, we took advantage of the differential placement of Ala150 in the structures of A6 bound to Tax and Tel1p: in the A6–Tel1p-HLA-A2 structure, Ala150 is at the N-terminus of a helical segment, whereas in the A6–Tax-HLA-A2 structure, Ala150 is at the C-terminus of a helical segment (Fig. 2E). As illustrated in Fig. 4A, mutagenesis of Ala150 to proline should therefore have opposing effects on the binding of A6 to the complexes of Tax or Tel1p with HLA-A2. For any position other than the N-terminus, proline destabilizes an helix as the i→i–4 hydrogen bond is lost and the carbons of the proline ring clash with the preceding backbone. At a helix N-terminus, however, there is no requirement for an i→i–4 hydrogen bond and there is no preceding helical geometry for the proline side-chain to interfere with. Also, proline is a strong helix initiator and has a high statistical preference for helix N-termini (Kim and Kang, 1999). We thus hypothesized that substitution of Ala150 with proline would 1) weaken TCR recognition of the Tax peptide as the helical structure necessary for binding is destabilized and 2) strengthen TCR recognition of the Tel1p peptide as the helical structure necessary for binding is stabilized.
Figure 4.
Proline mutagenesis verifies the switch that occurs in the HLA-A2 α2 helix upon A6 recognition of the Tel1p peptide and suggests coupling between motion in the α2 helix and the Tel1p peptide. A) Proline at position 150 would break the usual HLA-A2 α2 helix conformation, as a hydrogen bond is lost and interatomic bumps are introduced (left; additional atoms from the proline ring are yellow). In contrast, the altered conformation seen in A6 recognition of the Tel1p peptide can tolerate aproline at position 150 (right). B) Results for the A6 TCR binding wild type or A150P peptide-HLA-A2 presenting the Tax peptide (left) or the Tel1p peptide (right). Consistent with the structural data, the A150P mutation weakens affinity for Tax but enhances affinity for Tel1p. C) The structures of the A150P HLA-A2 mutant presenting the Tel1p and Tax peptide verify the mutation stabilizes the altered conformation. The “normal” α2 helix conformation from the unligated Tel1p complex is grey, the conformation from the A6–Tel1p-HLA-A2 ternary complex is cyan, the conformation from the unligated A150P mutant with Tel1p is brown, and the conformation from the unligated A150P mutant with Tax is burgundy. Superimposition is via backbone atoms of the peptide-binding domains. D) Electron density for peptide side chains in the A150P Tel1p structure (top) is weaker than the density in the A150P Tax structure (bottom), despite the fact that the Tel1p structure is of slightly better resolution. This suggests a coupling between mobility in the Tel1p peptide and mobility in the HLA-A2 α2 helix. Densities are 2Fo-Fc contoured at 1σ.
Figure 4B shows the results of Biacore binding experiments for A6 binding the Tax and Tel1p peptides presented by wild-type and A150P HLA-A2. As hypothesized, the A150P mutation weakens TCR affinity for Tax-HLA-A2, reducing the KD from 2 μM to 10 μM (equivalent to a ΔΔG° of 1.0 kcal/mol). Conversely, the mutation strengthens TCR affinity for Tel1p-HLA-A2, enhancing the KD from 41 μM to 5 μM (equivalent to a ΔΔG° of −1.2 kcal/mol). The differential effect of the A150P mutation independently verifies the crystallographically observed conformational switch in HLA-A2.
To confirm that the results with the A150P mutation did in fact report on the conformational properties of the HLA-A2 α2 helix and were not an artifact themselves, we determined the crystallographic structures of the A150P mutant of HLA-A2 presenting the Tel1p and the Tax peptides at 2.0 and 2.1 Å resolution, respectively (Table 1). As shown in Fig. 4C, the HLA-A2 α2 helix in these structures closely mimics the altered conformation seen in the A6–Tel1p-HLA-A2 ternary complex, confirming the conclusions from the binding studies (electron density images for the helices in these structures are in Supplemental Fig. S6)
Interestingly, in the A150P Tel1p-HLA-A2 structure, there was weaker electron density for the side chains of Tyr5 and Gln7 of the Tel1p peptide than in the structure of the wild-type Tel1p-HLA-A2 complex, despite the two structures being of similar resolution and quality (Fig. 4D, compare with Fig. S1). The electron density for Tyr5 was also weaker than that for Tyr5 in the A150P Tax-HLA-A2 structure, which crystallized in the same form as A150P Tel1p-HLA-A2 and even diffracted to a poorer resolution. The observation of weaker electron density for the Tel1p side chains when the HLA-A2 α2 helix is shifted suggests a coupling between the motional properties of the peptide and the α2 helix when Tel1p but not Tax is bound to HLA-A2 (there were no crystallographic contacts to the peptide or α2 helix linker region in either of the A150P peptide-HLA-A2 structures). In summary, the data with the A150P mutant confirms the shift in the α2 helix upon A6 recognition Tel1p-HLA-A2 and suggest a linkage between flexibility in the peptide and flexibility in HLA-A2 when Tel1p but not Tax is bound.
HLA-A2 possesses greater conformational dynamics when Tel1p is presented as opposed to Tax
Although the structural and binding data are reminiscent fan induced-fit mechanism for the conformational change in the HLA-A2 α 2 helix upon TCR recognition of the Tel1p peptide, a traditional induced-fit mechanism seems unlikely given the close similarities in the unligated Tax-HLA-A2 and Tel1p-HLA-A2 complexes (Fig. 1). Moreover, emerging data from other systems suggests that intrinsic dynamics in an unligated molecule can be responsible for conformational changes observed upon binding (James et al., 2003; Lange et al., 2008; Tobi and Bahar, 2005). To investigate the possibility that flexibility in the unligated Tel1p-HLA-A2 complex could contribute to the conformational changes seen upon TCR binding, we examined the dynamical properties of the Te1p-HLA-A2 α2 helix more closely.
First, we substituted His151 in the α2 linker region with cysteine. His151 is solvent exposed in the structures of HLA-A2 with Tax and Tel1p and does not contact any other HLA-A2 side-chains. The H151C HLA-A2 mutant was then refolded with either the Tax or Tel1p peptide and labeled at the free cysteine with fluorescein-5-maleimide. Following re-purification, the steady-state fluorescence anisotropy of the labeled Tax and Tel1p HLA-A2 complexes was measured. The anisotropy values of both complexes were in the range of 0.15 to 0.20, similar to the values reported for fluorescently-labeled peptides bound to HLA-A2 (Binz et al., 2003). However, as shown in Fig. 5A, the steady-state anisotropy of the Tel1p-HLA-A2 complex was 25% lower than that of Tax-HLA-A2. Several control experiments ensured that the difference in anisotropy between the Tel1p and Tax HLA-A2 complexes could not be attributed to non-specific labeling, unincorporated label, or differences in pMHC stability (see Experimental Procedures).
Figure 5.
The HLA-A2 α2 helix linker region is more dynamic when the Tel1p rather than the Tax peptide is presented. A) Steady-state anisotropies for Tel1p-HLA-A2 fluoresce in (Flc) labeled at position 151 are 25% lower than those for Tax-HLA-A2 labeled at position 151, indicating greater backbone flexibility in the Tel1p complex. The position 151 values are substantially higher than those for protein labeled at position 195, located in a loop at the tip of the α3 domain. Anisotropies for α2 helix positions 145 and 138, located outside of the linker region, are independent of which peptide is bound. Similar results were seen for position 151 when labeled with BODIPY-FL. Data shown are averages and standard deviations of 50 measurements performed at 25 °C on three independently generated samples at 100 nM. B) The differential dynamics of the HLA-A2 α2 helix revealed by steady-state anisotropy can be attributed to increased fluctuations occurring over the nanosecond timescale. In a time resolved experiment, 50% of the initial anisotropy at position 151 was lost by 8.4 ns for the Tax complex and 3.5 ns for the Tel1p complex. For comparison, position 195 lost 50% of its initial anisotropy in 1.1 ns.
Loss of anisotropy can occur via three mechanisms: global tumbling of the protein, rotation of the fluorescent core around its tether arm, and flexibility of the backbone to which the label is conjugated. As protein tumbling and rotation of the dye will be the same in the labeled Tax-HLA-A2 and Tel1p-HLA-A2 complexes, the lower anisotropy value for the Tel1p complex indicates greater dynamics for the α2 helix backbone in the Tel1p complex compared to the Tax complex.
To ensure our measurements were properly differentiating between different degrees of dynamics, we examined a highly flexible region of HLA-A2. The backbone of Ser195, in a distal loop at the tip of the HLA-A2 α3 domain is occasionally disordered in structures with class I pMHC complexes (e.g., Gagnon et al., 2006). Consistent with this position possessing high intrinsic dynamics, when labeled with fluorescein, position 195 exhibited a low anisotropy independent of which peptide was bound (Fig. 5A). In addition to serving as a positive control for the dynamics measurements, this observation helps explain why dynamical differences in the α2 helix linker region were not evident from comparisons of the Tel1p-HLA-A2 and Tax-HLA-A2 crystal structures: although the α2 helix linker region is more dynamic when the Tel1p peptide is bound, it does not possess the high degree of dynamics characteristic of flexible loops.
We next examined positions in the peptide binding domain that are not structurally altered upon A6 recognition of either Tel1p-HLA-A2 or Tax-HLA-A2. His145 is with in the short arm of the α2 helix and Met138 is at its N-terminal end. The side chains of both amino acids are solvent exposed and do not contact atoms of the peptide or HLA-A2 in the unligated Tax-HLA-A2 or Tel1p-HLA-A2 structures. When cysteine was introduced at these positions and labeled with fluoresce in, the anisotropy was independent of which peptide was bound (Fig. 5A), indicating that the peptide-dependent dynamical differences in HLA-A2 are localized to the region of the α2 helix that changes upon A6 recognition of the Tel1p ligand.
Lastly, we repeated the measurements at position 151 using BODIPY-FL, a fluorescent tag with a structure different from that of fluorescein. Again, the anisotropy with the Tel1p peptide was greater than with the Tax peptide (Fig. 5A). The absolute anisotropy values and the percentage difference between Tel1p and Tax were lower than observed with fluoresce in, attributable to BODIPY-FL’s longer fluorescent lifetime (6 ns vs. 4 ns) and longer tether arm (two additional rotatable bonds), which allows overall protein tumbling and dye rotation to have a greater impact in fluorescence depolarization.
To complement the steady-state fluorescence anisotropy measurements, we next examined the time decay of anisotropy using single photon counting time-resolved fluorescence anisotropy (TRFA). In contrast with the steady-state measurements which measured the anisotropy under constant illumination, this experiment followed the decay of anisotropy following a rapid pulse. The advantage of TRFA is that information regarding motional timescales can be collected. As shown in Fig. 5B, with the fluorescein-labeled samples, the anisotropy decay in the Tel1p sample was more rapid with Tel1p than Tax, with 50% of the anisotropy lost by 3.5ns for Tel1p, compared to 8.4ns for Tax. The position 195 control had a much more rapid decay rate, with 50% of the anisotropy decay occurring with in 1.1ns. In summary, both the steady state and time resolved fluorescence anisotropy experiments reveal that the HLA-A2 α2 helix linker region is more mobile when the Tel1p rather than the Tax peptide is bound.
Free Tel1p-HLA-A2 samples its TCR-bound conformation and possesses coupled peptide-MHC flexibility
To help interpret the dynamics measurements, we next performed unrestrained molecular dynamics (MD) simulations on the unligated Tax-HLA-A2 and Tel1p-HLA-A2 complexes. Over the course of 50 ns of simulation time in explicit solvent, there were clear differences between the Tax and Tel1p complexes. In the Tel1p complex, amino acids in the α2 helix linker region sampled a range of φ,ψ bond angles, including those observed in the TCR-bound state (Fig. 6A). This was most noticeable for Ala149, which in the Tel1p simulation sampled both the TCR-bound and TCR-free conformations. In contrast, the region in the Tax simulation was more static. Examining Tyr5 and Gln7 of the Tel1p peptide, these side-chains were also found to populate both their TCR-bound and TCR-free conformations (Fig. 6B). These results could not be attributed to peptide dissociation or unfolding of the peptide binding groove, as in each simulation the anchoring hydrogen bonds between the heavy chain and the peptide primary anchors were retained.
Figure 6.
Molecular dynamics simulations of free pMHC complexes confirm the greater dynamics present in the Tel1p-HLA-A2 complex. A) Amino acids in the α2 helix linker region sample a range of ψ, φ bond angles in the Tel1p simulation, including both the TCR-free and TCR-bound conformations for Ala149. These positions in the Tax simulation are more static. B) Tyr5 and Gln7 sample both TCR-free and TCR-bound conformations in the Tel1p simulation. C) In the Tel1p simulation, the dynamic cross-correlation matrix reveals correlated motion between the C-terminal half of the peptide and the HLA-A2 α2 helix linker region. Little or no correlations are seen between the peptide and α2 helix in the Tax simulation.
We next computed dynamic cross-correlation matrices (DCCM) for the Tax and Tel1p simulations. The DCCM is composed of coefficients (Cij) describing the time-correlation of motion between components i and j (Ichiye and Karplus, 1991), in this case the carbons of the indicated residues. Cij values range from +1 to −1, with positive values reflecting correlated movement in the same direction and negative values reflecting correlated movement in the opposite direction. We focused the analysis on the peptide and the HLA-A2 α2 helix linker region as the structural data suggested coupled movement between these two components in Tel1p-HLA-A2 but not Tax-HLA-A2 (see Fig. 4D). In the Tel1p simulation, the analysis indicated that motion in Tyr5, Leu6, and Gln7 of the peptide were positively correlated to as much as 50% with motion in residues 149–155 of the α 2 helix (Fig. 6D). In contrast, there was little or no correlated movement between these regions for the Tax simulation. The DCCM data therefore revealed a path of dynamic communication between the peptide and α 2 helix in the Tel1p-HLA-A2 complex but not the Tax-HLA-A2 complex.
Tel1p-HLA-A2 is of low thermal stability and is recognized by A6 with disparate thermodynamics
A prediction from the results of the dynamics experiments with the Tax and Tel1p complexes with HLA-A2 is that the Tel1p-HLA-A2 complex should be of lower stability than the Tax-HLA-A2 complex. We thus examined the stability of both complexes using circular dichroism, monitoring the unfolding of the HLA-A2 molecule (Supplemental Fig. S7). The apparent Tm of the Tax-HLA-A2 complex was found to be 64 °C, in agreement with previous work (Khan et al., 2000). In contrast, the Tm of the Tel1p-HLA-A2 complex was lower at 55°C. As the Tel1p peptide shares optimal leucine and valine anchor residues with the Tax peptide, has fully occupied P1, P2, and P9 pockets, and adopts the usual extended conformation in the HLA-A2 peptide binding groove, the lower thermal stability of the Tel1p complex is best attributed to the greater dynamic instability of the pMHC complex.
As a final experiment to probe the difference between A6 recognition of the Tax and Tel1p ligands, we determined the thermodynamics for A6 binding Tel1p-HLA-A2 (Supplemental Fig. S8). The values for A6 recognition of Tax-HLA-A2 were determined previously (Davis-Harrison et al., 2005). Under the same conditions (HEPES buffer, pH 7.4, 25 °C), recognition of Tel1p-HLA-A2 is both enthalpically and entropically less favorable than recognition of Tax-HLA-A2(ΔH° of −3.0kcal/mol for Tel1p vs. −4.2 kcal/mol for Tax, ΔS° of 9cal/mol/K for Tel1p vs. 12 for Tax). Although different results should be expected from the two interfaces (Armstrong et al., 2008a), the data are consistent with both the structural and dynamical observations: the unfavorable shift in enthalpy agrees with the need to stabilize alternate conformations of the Tel1p peptide and HLA-A2 α2 helix, and the unfavorable shift in entropy agrees with the binding-associated reduction of conformational dynamics present in Tel1p-HLA-A2 but not Tax-HLA-A2.
Discussion
Despite serving as a sequence and structural mimic for Tax-HLA-A2, the Tel1p-HLA-A2 complex forms an unexpectedly different interface with the A6 TCR, with conformational differences in the peptide, the HLA-A2 molecule, and the receptor. The observation is in contrast to other examples of a single TCR recognizing different peptides presented by the same MHC molecule (Degano et al., 2000; Garcia et al., 1998; Reiser et al., 2000; Reiser et al., 2003), and is inconsistent with a molecular mimicry mechanism as traditionally envisioned.
Why is recognition of Tel1p so much more complex compared to recognition of Tax? A conventional explanation would focus on the small number of steric clashes that occur when the structure of unligated Tel1p-HLA-A2 is superimposed onto Tax-HLA-A2 in the A6–Tax-HLA-A2 structure (Fig. S4). Yet these clashes are not extensive and can be remedied by small shifts in side chain positions. A more crucial observation is that the unbound Tel1p-HLA-A2 molecule is more dynamic than the Tax-HLA-A2 molecule, and that in Tel1p-HLA-A2, both the peptide and the HLA-A2 α 2 helix at least partially sample their TCR-bound conformations. These findings suggest a conformational selection mechanism, whereby the TCR interacts preferentially with an alternate, binding-competent conformation of the pMHC as it is sampled (Ma et al., 1999). Conformational selection is consistent with much emerging data on how flexibility can influence the specificity of protein-protein interactions (James et al., 2003; Lange et al., 2008; Tobi and Bahar, 2005). Alternatively, as enhanced dynamics result from reduced energy barriers separating different protein conformations, the conformational sampling in the unbound molecule could facilitate induced-fit-type changes in the peptide and/or HLA-A2 α2 helix that occur as binding proceeds. In either case though, the picture that emerges is that the ability of A6 to recognize both the Tax and Tel1p ligands depends at least in part upon the differing degree of mobility present within the two-pMHC complexes.
The influence of peptide flexibility in TCR recognition of pMHC has been discussed previously. Most related to the results presented here is the notion that differential peptide flexibility resulting from MHC polymorphisms can broaden TCR reactivity (Arch bold et al., 2009; Fabian et al., 2008; Pohlmann et al., 2004). Our findings extend this concept by showing that not only can different peptides possess different dynamic character; they can affect the dynamics of the MHC peptide-binding groove in a way that facilitates TCR engagement. Thus TCR recognition, and by extension cross-reactivity, can be directed by the dynamical character imparted on the ligand by different peptides.
If differing degrees of ligand flexibility can explain the structural differences in the peptide and HLA-A2 molecule seen upon A6 recognition of Tax and Tel1p, what mechanism explains the different TCR conformations in the two TCR-pMHC interfaces? The structure of the A6–Tel1p-HLA-A2 complex is the sixth ternary complex with A6 showing a different conformation for CDR3β (Ding et al., 1999; Gagnon et al., 2006; Garboczi et al., 1996; Piepen brink et al., 2009), suggesting that the CDR3β loop possesses its own degree of dynamic instability. This is further supported by a wealth of binding and mutational data with A6 (Armstrong and Baker, 2007; Davis-Harrison et al., 2005; Laugel et al., 2005), as well as the general observation that CDR3 loops in TCRs tend to adopt different conformations free and bound (Armstrong et al., 2008b). It is thus possible that the conformation of CDR3β seen in recognition of Tel1p is of equal or even lower energy than that the conformation seen in recognition of Tax. If this is the case, upon recognition of Tel1p, CDR3β could adopt a more favorable conformation at the expense of a less favorable Tel1p-HLA-A2 conformation. This option would not be available for recognition of Tax due to the greater rigidity of the Tax-HLA-A2 complex, forcing the TCR to adopt a less favorable conformation in order to optimize packing. This would not fully explain why Tel1p is recognized more weakly than Tax, but other differences between the two interfaces (such as hydrogen bonding and hydrophobic contacts) will also influence affinity. Testing this hypothesis, which essentially describes TCR-pMHC recognition as the cooperative “melding” of two conformational ensembles, will require more detailed studies of the flexibility of the TCR CDR loops.
Lastly, why is the Tel1p-HLA-A2 complex more dynamic than the Tax-HLA-A2 complex? Identifying the origins of protein dynamics from crystallographic structures remains a challenge. Yet one clue lies in the correlation between movement in the Tel1p peptide and movement in the HLA-A2 α2 helix observed both crystallographic ally and in the molecular simulations. One interpretation consistent with the data is that rotation of Tyr5 due to thermal fluctuations influences rotation of Gln7 (and vice versa), which together influence the positions of adjacent positions in the HLA-A2 α2 helix.
The interpretation above would also explain why the Tax complex is more rigid than the Tel1p complex. In the unbound state of either pMHC complex, rotation of the Tyr5 side-chain cannot occur unless other elements of the peptide move out of the way. As illustrated in Fig. 2C, in the Tel1p complex this occurs via rotation of the Tyr5 ψ bond, followed by a counter-rotation of the Leu6 φ bond to keep the path of the peptide backbone intact. In the Tax complex, rotation is prevented by the presence of proline rather than leucine at position 6. Thus the Tax peptide, and by extension the HLA-A2 α2 helix, remains locked into a smaller conformational ensemble. This would also explain why observations similar to ours have not yet been made in other TCR-pMHC systems, as the relationship between peptide and MHC dynamics will necessarily be peptide specific. The underlying phenomena, that the conformational ensemble of a protein can be differentially “tuned” by various binding partners has been suggested in theoretical studies of protein dynamics and is compatible with the energy landscape model of protein conformation and dynamics (Ma et al., 1999; Miller and Dill, 1997).
In conclusion, we have demonstrated that the ability of the A6 TCR to recognize both the Tax and Tel1p ligands hinges on the differing degrees of molecular motion present in the two peptides and the HLA-A2 α2 helix. These results shed new light on how structural diversity can be presented to and accommodated by receptors of the immune system, and reveal that antigen-dependent tuning of molecular motion distributed throughout the TCR binding surface of the pMHC molecule can contribute to TCR recognition and facilitate cross-reactivity.
Experimental Procedures
Proteins and peptides
HLA-A2 and the A6 TCR were refolded from bacterially expressed inclusion bodies and chromatographically purified as previously described (Davis-Harrison et al., 2005). Peptides were synthesized locally on an ABI433A instrument; purity and mass were verified by LC-MS. The TCR construct used included an engineered disulfide bond linking the two constant domains (Boulter et al., 2003).
X-ray crystallography
Peptide-HLA-A2 crystals were grown from 24% PEG 3350 in 25 mM MES, pH 6.5, 0. 1 M NaF. A6– Tel1p-HLA-A2 crystals were grown in 15% PEG 4000 in 0.1 M Tris, pH 8.5, 0.2 M MgCl2. Cryo-protection consisted of 20–25% glycerol. Diffraction data were collected at Argonne National Laboratory at the indicated beam lines. Data reduction, structure solution, refinement, and structure validation was performed as previously described (Gagnon et al., 2006), with the addition of composite omit maps to evaluate positions in the models. Composite omit maps, calculated in CNX (Accelrys), iteratively excluded 5% of the mode land included simulated annealing. Search models for molecular replacement were PDB entries 1 TVB for pMHC (Borbulevych et al., 2005) and 2GJ6 for TCR-pMHC (Gagnon et al., 2006).
Surface plasmon resonance
Binding experiments were performed using Biacore 3000 and T100 surface plasmon resonance instruments with the TCR on the sensor surface as previously described (Davis-Harrison et al., 2005). Measurements were performed in 20 mM HEPES, 150 mM NaCl, 0.005% P-20, pH 7.4. All affinities were measured using steady-state binding measurements, and each injection was repeated twice. Thermodynamic measurements were performed as previously described (Cole et al., 2009).
Fluorescence anisotropy
For fluorescent labeling, various amino acids in the HLA-A2 α2 helix linker region were replaced with cysteine. Following normal refolding and purification, the cysteine mutant pMHC molecules were labeled with fluorescein-5-maleimide or BODIPY-FL N-(2-aminoethyl)-maleimide at room temperature for one hour. To remove free dye, proteins were first extensively dialyzed against 1000-fold buffer excess for 24 hours with three buffer changes. Dialyzed proteins were then re-purified via size-exclusion chromatography, followed by a second round of dialysis. Performing side-by-side reactions with wild-type pMHC controlled all labeling reactions. Nonspecific labeling never reached above background fluorescence, whereas specific labeling for cysteine mutants ranged from 70% to 90%. Steady-state fluorescence anisotropy measurements were performed using a Beacon 2000 instrument (Invitrogen). Sample conditions were 20 mM sodium phosphate; 75 mM NaCl, pH 7.4. Labeling, purification, and measurements were repeated three times with freshly prepared protein. Anisotropy reading son individual samples were repeated 50 times. The difference in anisotropies between the samples labeled with fluoresce in at position 151 persisted at 4 °C, 25 °C, and 37 °C, persisted in the presence of 20-fold excess peptide, and was independent of concentration, from 10 nM – 50 μM. Anisotropy values on both complexes also remained constant during extended incubations at room temperature (up to 48 hours). As a final control to verify the peptide-dependence of the anisotropy and ensure free dye did not contribute to the measured values, purified Tel1p-HLA-A2 fluorescein-labeled at position 151 was unfolded in 8M urea, pH 10, separated from free peptide by exhaustive dialysis against denaturant, and the labeled heavy chain recovered and refolded with the Tax peptide and β2m per the usual procedure. The difference in anisotropy measured in this experiment was identical to that determined from separately labeled Tel1p-HLA-A2 and Tax-HLA-A2 (see Supplemental Table S1 for a summary of these control experiments).
Time resolved measurements were performed with a FluoroCube 5000U time correlated single photon counting fluorimeter (HORIBA Jobin Yvon) using a 457 nm NanoLED pulsed excitation source. Sample conditions were the same as the steady-state measurements, with pMHC concentrations of 3μM. Experiments were performed at 4 °C. For each sample, five sets of measurements were collected that included alternating parallel and perpendicular intensity readings at 30-second intervals; all data were included in the subsequent sum/difference anisotropy analysis. Data were processed and analyzed with the DAS6 software distributed with the instrument.
Circular dichrosim
Circular dichrosim measurements of thermal stability were performed using an Aviv 62DS spectrometer monitoring 218 nm as previously described (Khan et al., 2000). Solution conditions were 20 mM phosphate and 75 mM NaCl (pH 7.4). Protein concentrations were 10 μM. A temperature increment of approximately 0.3°C/min was used. Because unfolding is irreversible, data were fit to a six-order polynomial and the apparent Tm taken from the first derivative of the fitted curve.
Molecular dynamics simulations
MD simulations for the Tel1p and Tax complexes with HLA-A2 were performed using the AMBER9 package (Case et al., 2005). Starting coordinates were from the second molecule in each asymmetric unit. The systems were solvated with a truncated octahedral box of explicit TIP3P water that extended a minimum of 12 Å from the solute atoms; crystallographic waters were kept if they were 3.5 Å or closer to hydrogen bond donors or acceptors. Sodium cations were added for neutrality. For equilibration, first the proteins were constrained while surrounding water was optimized. Constraints were then gradually removed and the entire systems minimized unrestrained. The systems were then heated to the production temperature over two 50 ps dynamics runs at constant volume, followed by 1.1ns of equilibration and then 50 ns of production at constant pressure. Because the anisotropy experiments indicated an approximately 25% difference in dynamics, we used a slightly elevated production temperature of 330K to enhance sampling. Temperature was controlled using the Lange in scheme. The SHAKE algorithm was used, allowing a 2 fs time step. Dynamic cross-correlation matrices (DCCM) were computed with ptraj and visualized with Matlab. Superimpositions for DCCM calculations were via the backbones of the α1-α2 helices and the peptides.
Accession Codes
PDB accession codes for the structures determined in this study are 3H7B for Tel1p-HLA-A2, 3H9S for A6–Tel1p-HLA-A2, 3H9H for A150P Tel1p-HLA-A2, and 3IXA for A150P Tax-HLA-A2.
Supplementary Material
Acknowledgments
We thank F. Insaidoo for helpful discussion, A. Wojnarowicz for technical assistance, and P. Kamat for use of a FluoroCube 5000U instrument. Supported by grants R01GM067079 from NIGMS, NIH and MCB0448298 from NSF. Results shown in this report are derived from work performed at the Structural Biology Center and Lilly Research Laboratories Collaborative Access Teams (SBC-CAT and LRL-CAT) at the Advanced Photon Source, Argonne National Laboratory. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy under contract DE-AC02-06CH11357. Eli Lilly & Company who operates the facility provided use of the LRL-CAT facilities at Sector 31 of the Advanced Photon Source. The Notre Dame CBBI training program, funded by T32GM075762 from NIGMS, NIH, supported KHP.
References
- Arch bold JK, Macdonald WA, Gras S, Ely LK, Miles JJ, Bell MJ, Brennan RM, Beddoe T, Wilce MCJ, Clements CS, et al. Natural micropolymorphism in human leukocyte antigens provides a basis for genetic control of antigen recognition. J Exp Med. 2009;206:209–219. doi: 10.1084/jem.20082136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong KM, Baker BM. A Comprehensive Calorimetric Investigation of an Entropically Driven T Cell Receptor-Peptide/Major Histocompatibility Complex Interaction. Biophys J. 2007;93:597–609. doi: 10.1529/biophysj.107.104570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong KM, Insaidoo FK, Baker BM. Thermodynamics of T-cell receptor-peptide/MHC interactions: progress and opportunities. Journal of Molecular Recognition. 2008a;21:275–287. doi: 10.1002/jmr.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong KM, Piepenbrink KH, Baker BM. Conformational changes and flexibility in T-cell receptor recognition of peptide-MHC complexes. Biochem J. 2008b;415:183–196. doi: 10.1042/BJ20080850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binz AK, Rodriguez RC, Biddison WE, Baker BM. Thermodynamic and kinetic analysis of a peptide-class I MHC interaction highlights the noncovalent nature and conformational dynamics of the class I heterotrimer. Biochemistry. 2003;42:4954–4961. doi: 10.1021/bi034077m. [DOI] [PubMed] [Google Scholar]
- Borbulevych OY, Baxter TK, Yu Z, Restifo NP, Baker BM. Increased Immunogenicity of an Anchor-Modified Tumor-Associated Antigen Is Due to the Enhanced Stability of the Peptide/MHC Complex: Implications for Vaccine Design. J Immunol. 2005;174:4812–4820. doi: 10.4049/jimmunol.174.8.4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulter JM, Glick M, Todorov PT, Baston E, Sami M, Rizkallah P, Jakobsen BK. Stable, soluble T-cell receptor molecules for crystallization and therapeutics. Protein Eng. 2003;16:707–711. doi: 10.1093/protein/gzg087. [DOI] [PubMed] [Google Scholar]
- Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. Journal of Computational Chemistry. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements CS, Dunstone MA, Macdonald WA, McCluskey J, Rossjohn J. Specificity on a knife-edge: the [alpha][beta] T cell receptor. Current Opinion in Structural Biology. 2006;16:787–795. doi: 10.1016/j.sbi.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Cole DK, Yuan F, Rizkallah PJ, Miles JJ, Gostick E, Price DA, Gao GF, Jakobsen BK, Sewell AK. Germline-governed recognition of a cancer epitope by an immunodominant human T-cell receptor. Journal of Biological Chemistry. 2009 doi: 10.1074/jbc.M109.022509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colf LA, Bankovich AJ, Hanick NA, Bowerman NA, Jones LL, Kranz DM, Garcia KC. How a Single T Cell Receptor Recognizes Both Self and Foreign MHC. Cell. 2007;129:135–146. doi: 10.1016/j.cell.2007.01.048. [DOI] [PubMed] [Google Scholar]
- Davis-Harrison RL, Armstrong KM, Baker BM. Two Different T Cell Receptors use Different Thermodynamic Strategies to Recognize the Same Peptide/MHC Ligand. Journal of Molecular Biology. 2005;346:533–550. doi: 10.1016/j.jmb.2004.11.063. [DOI] [PubMed] [Google Scholar]
- Degano M, Garcia KC, Apostolopoulos V, Rudolph MG, Teyton L, Wilson IA. A functional hot spot for antigen recognition in a superagonist TCR/MHC complex. Immunity. 2000;12:251–261. doi: 10.1016/s1074-7613(00)80178-8. [DOI] [PubMed] [Google Scholar]
- Ding YH, Baker BM, Garboczi DN, Biddison WE, Wiley DC. Four A6-TCR/peptide/HLA-A2 structures that generate very different T cell signals are nearly identical. Immunity. 1999;11:45–56. doi: 10.1016/s1074-7613(00)80080-1. [DOI] [PubMed] [Google Scholar]
- Fabian H, Huser H, Narzi D, Misselwitz R, Loll B, Ziegler A, Bockmann RA, Uchanska-Ziegler B, Naumann D. HLA-B27 subtypes differentially associated with disease exhibit conformational differences in solution. Journal of Molecular Biology. 2008;376:798–810. doi: 10.1016/j.jmb.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Gagnon SJ, Borbulevych OY, Davis-Harrison RL, Turner RV, Damirjian M, Wojnarowicz A, Biddison WE, Baker BM. T Cell Receptor Recognition via Cooperative Conformational Plasticity. Journal of Molecular Biology. 2006;363:228–243. doi: 10.1016/j.jmb.2006.08.045. [DOI] [PubMed] [Google Scholar]
- Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- Garcia KC, Degano M, Pease LR, Huang M, Peterson PA, Teyton L, Wilson IA. Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen. Science. 1998;279:1166–1172. doi: 10.1126/science.279.5354.1166. [DOI] [PubMed] [Google Scholar]
- Hausmann S, Biddison WE, Smith KJ, Ding YH, Garboczi DN, Utz U, Wiley DC, Wucherpfennig KW. Peptide recognition by two HLA-A2/Tax11-19-specific T cell clones in relationship to their MHC/peptide/TCR crystal structures. J Immunol. 1999;162:5389–5397. [PubMed] [Google Scholar]
- Ichiye T, Karplus M. Collective motions in proteins: A covariance analysis of atomic fluctuations in molecular dynamics and normal mode simulations. Proteins: Structure, Function, and Genetics. 1991;11:205–217. doi: 10.1002/prot.340110305. [DOI] [PubMed] [Google Scholar]
- Ishizuka J, Stewart-Jones GBE, van der Merwe A, Bell JI, McMichael AJ, Jones EY. The Structural Dynamics and Energetics of an Immunodominant T Cell Receptor Are Programmed by Its V[beta] Domain. Immunity. 2008;28:171–182. doi: 10.1016/j.immuni.2007.12.018. [DOI] [PubMed] [Google Scholar]
- James LC, Roversi P, Tawfik DS. Antibody Multispecificity Mediated by Conformational Diversity. Science. 2003;299:1362–1367. doi: 10.1126/science.1079731. [DOI] [PubMed] [Google Scholar]
- Khan AR, Baker BM, Ghosh P, Biddison WE, Wiley DC. The structure and stability of an HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site. J Immunol. 2000;164:6398–6405. doi: 10.4049/jimmunol.164.12.6398. [DOI] [PubMed] [Google Scholar]
- Kim MK, Kang YK. Positional preference of proline in alpha-helices. Protein Sci. 1999;8:1492–1499. doi: 10.1110/ps.8.7.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohm AP, Fuller KG, Miller SD. Mimicking the way to autoimmunity: an evolving theory of sequence and structural homology. Trends in Microbiology. 2003;11:101–105. doi: 10.1016/s0966-842x(03)00006-4. [DOI] [PubMed] [Google Scholar]
- Lange OF, Lakomek NA, Fares C, Schroder GF, Walter KF, Becker S, Meiler J, Grubmuller H, Griesinger C, de Groot BL. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science. 2008;320:1471–1475. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- Laugel B, Boulter JM, Lissin N, Vuidepot A, Li Y, Gostick E, Crotty LE, Douek DC, Hemelaar J, Price DA, et al. Design of Soluble Recombinant T Cell Receptors for Antigen Targeting and T Cell Inhibition. J Biol Chem. 2005;280:1882–1892. doi: 10.1074/jbc.M409427200. [DOI] [PubMed] [Google Scholar]
- Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- Ma B, Kumar S, Tsai CJ, Nussinov R. Folding funnels and binding mechanisms. Protein Eng. 1999;12:713–720. doi: 10.1093/protein/12.9.713. [DOI] [PubMed] [Google Scholar]
- Mazza C, Auphan-Anezin N, Gregoire C, Guimezanes A, Kellenberger C, Roussel A, Kearney A, van der Merwe PA, Schmitt-Verhulst AM, Malissen B. How much can a T-cell antigen receptor adapt to structurally distinct antigenic peptides? EMBO J. 2007;26:1972–1983. doi: 10.1038/sj.emboj.7601605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DW, Dill KA. Ligand binding to proteins: The binding landscape model. Protein Sci. 1997;6:2166–2179. doi: 10.1002/pro.5560061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenbrink KH, Borbulevych OY, Sommese RF, Clemens J, Armstrong KM, Desmond C, Do P, Baker BM. Fluorine substitutions in an antigenic peptide selectively modulate T-cell receptor binding in a minimally perturbing manner. Biochemical Journal. 2009;423:353–361. doi: 10.1042/BJ20090732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlmann T, Bockmann RA, Grubmuller H, Uchanska-Ziegler B, Ziegler A, Alexiev U. Differential peptide dynamics is linked to MHC polymorphism. J Biol Chem. 2004;279:28197–28201. doi: 10.1074/jbc.C400128200. [DOI] [PubMed] [Google Scholar]
- Reiser JB, Darnault C, Guimezanes A, Grégoire C, Mosser T, Schmitt-Verhulst AM, Fontecilla-Camps JC, Malissen B, Housset D, Mazza G. Crystal structure of a T cell receptor bound to an allogeneic MHC molecule. Nat Immunol. 2000;1:291–297. doi: 10.1038/79728. [DOI] [PubMed] [Google Scholar]
- Reiser JB, Darnault C, Gregoire C, Mosser T, Mazza G, Kearney A, van der Merwe PA, Fontecilla-Camps JC, Housset D, Malissen B. CDR3 loop flexibility contributes to the degeneracy of TCR recognition. Nat Immunol. 2003;4:241–247. doi: 10.1038/ni891. [DOI] [PubMed] [Google Scholar]
- Rudolph MG, Speir JA, Brunmark A, Mattsson N, Jackson MR, Peterson PA, Teyton L, Wilson IA. The crystal structures of K(bm1) and K(bm8) reveal that subtle changes in the peptide environment impact thermostability and alloreactivity. Immunity. 2001;14:231–242. doi: 10.1016/s1074-7613(01)00105-4. [DOI] [PubMed] [Google Scholar]
- Smock RG, Gierasch LM. Sending Signals Dynamically. Science. 2009;324:198–203. doi: 10.1126/science.1169377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobi D, Bahar I. Structural changes involved in protein binding correlate with intrinsic motions of proteins in the unbound state. Proceedings of the National Academy of Sciences. 2005;102:18908–18913. doi: 10.1073/pnas.0507603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tynan FE, Reid HH, Kjer-Nielsen L, Miles JJ, Wilce MCJ, Kostenko L, Borg NA, Williamson NA, Beddoe T, Purcell AW, et al. A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat Immunol. 2007;8:268–276. doi: 10.1038/ni1432. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.