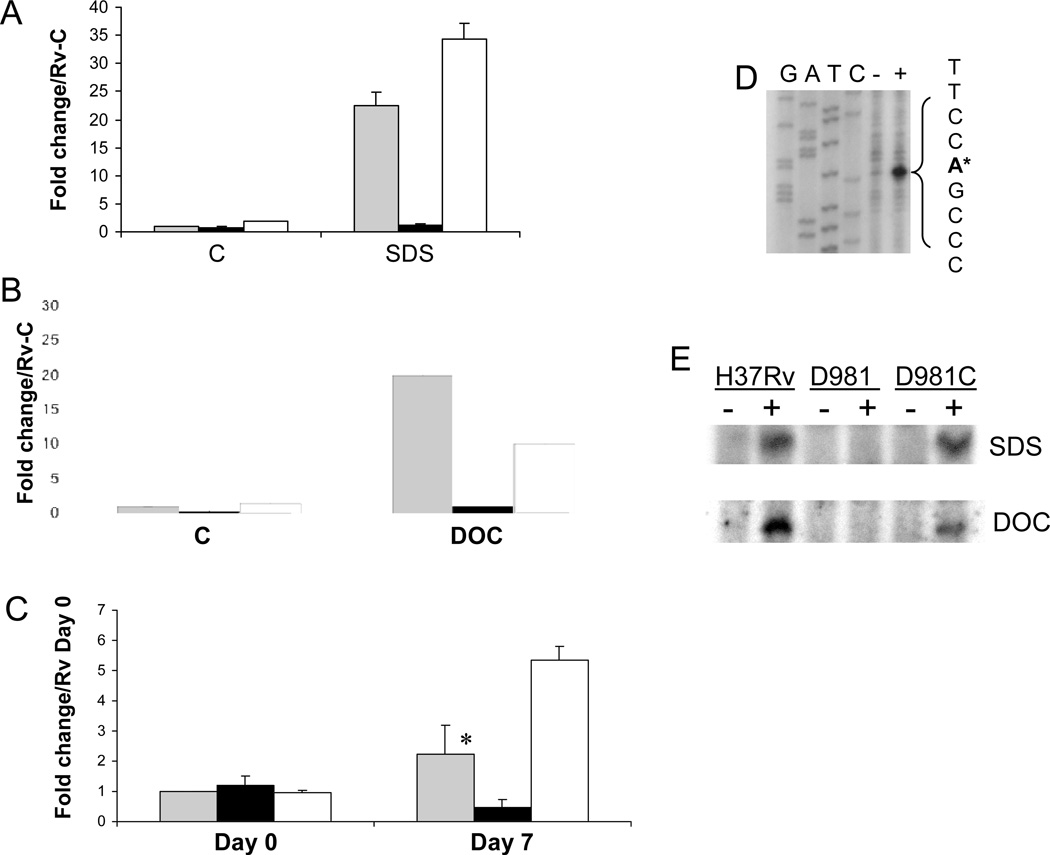
Figure 1.
MprAB activates Rv1057 under envelope stress. (A–C) Expression of Rv1057 was examined by qRT-PCR using RNA extracted from strains exposed to 0.05% SDS for 90 min (A), 0.1% deoxycholate for 90 min (B), or following infection of THP-1 cells (C). H37Rv (gray bars), Rv-D981 (black bars), and Rv-D981Com (white bars). The “C” on the horizontal axis in (A) and (B) indicates control (untreated) samples. For (C), activated THP-1 cells were infected at a MOI of 1:1, and bacteria were recovered from lysed monolayers at 2 hr (Day 0), or at day 7. Results are shown as fold change over the H37Rv control sample for panels A and B, or over the H37Rv Day 0 sample for panel C. Control and day 0 samples were given values of 1. For all panels, results were normalized for 16S RNA content and show the mean of at least two separate experiments +/− SD. (C) The difference in expression between H37Rv and Rv-D981 at day 7 was analyzed using a Student’s one-tailed T-test, *, P < 0.05.
(D) Rv1057 TSP2 was mapped by primer extension with primer Rv1057 Ext-3, using RNA extracted from H37Rv incubated in the absence (−) or presence (+) of 0.05% SDS. The TSP is marked by an asterisk and was located using a sequence ladder generated with the primer used for primer extension. The GATC labels above the ladder refer to the sequence on the opposite DNA strand. (E) Activation of Rv1057 TSP was compared in H37Rv, Rv-D981 (D981), and Rv-D981Com (D981C) by primer extension. Minus sign, cultures incubated under control conditions. Plus sign, cultures exposed to 0.05 % SDS (upper panel), or 0.1% deoxycholate (lower panel) for 90 min.