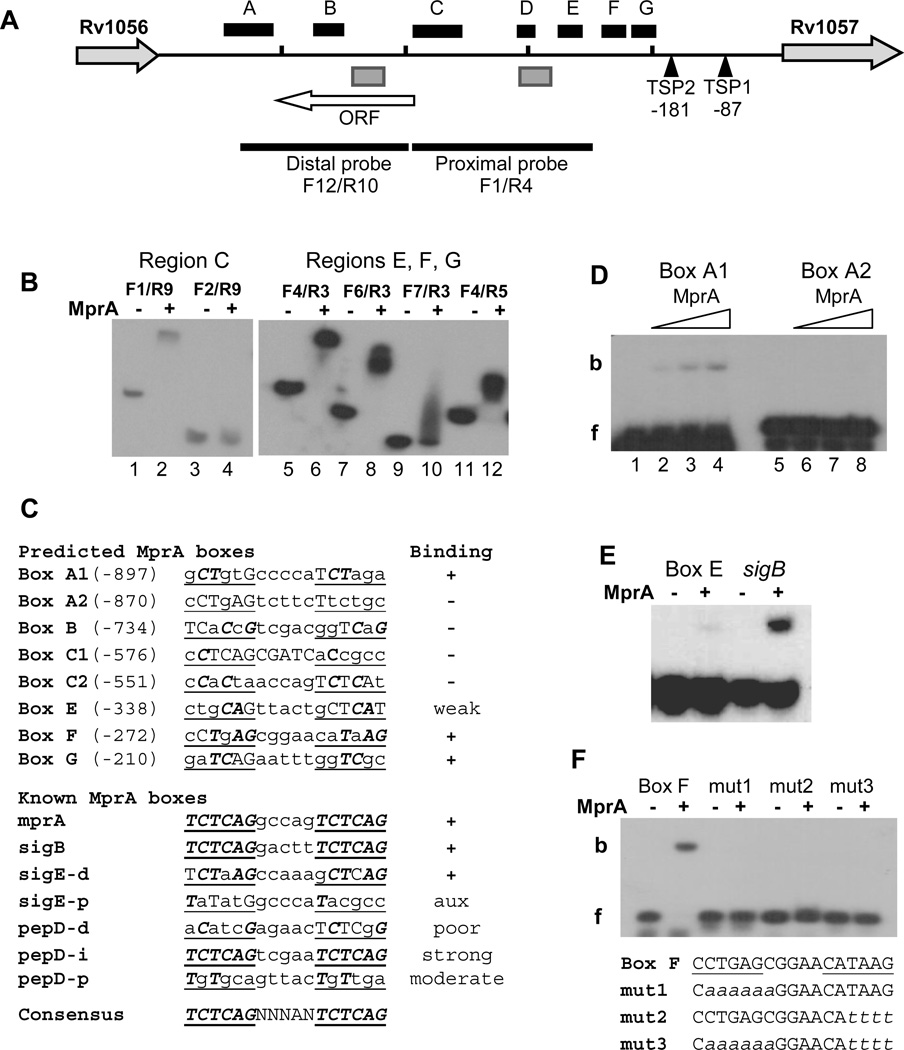
Figure 3.
Localization of MprA boxes in the Rv1056-Rv1057 intergenic region. (A) Features of the Rv1056-Rv1057 intergenic region. The horizontal black line represents the chromosome and short vertical bars mark 200-bp sections beginning from the Rv1057 start codon. The location of TSP1 30 and TSP2 (this study) are indicated relative to the Rv1057 start codon. Black boxes labeled A–G designate regions determined to contain at least one MprA box, based on EMSA analyses of overlapping fragments. Gray boxes designate regions containing TrcR binding sites 30 (and this study). The locations of the probes used in Figures 2 A and B are marked by thick black lines. The white horizontal arrow indicates the location and length of a previously unidentified ORF. Rv1057 and Rv1056 are not drawn to scale.
(B) Representative EMSAs used to localize MprA binding in region C (left panel) and regions E, F, and G (right panel). Forward and reverse primers used to generate each probe are noted. (−) and (+) symbols indicate incubation in the absence, or presence, of 2 ug MprA, respectively. Results indicate strong MprA binding to probes F1/R9, F4/R3, F6/R3, and F4/R5.
(C) Comparison of predicted MprA boxes in regions A–G with known MprA boxes. Regions were scanned for sequences with strongest similarity to the published consensus sequence for the MprA box 25, 26, 28. Repeated hexamers of the MprA box half-sites are underlined, bases matching the consensus sequences are capitalized, and capitals in bold italics mark consensus sequence bases present in the same position in each half-site. Names of predicted boxes are derived from the region in which they were found. Two boxes were predicted for each of regions A and C. Numbers in parentheses refer to the position of the first base relative to the Rv1057 start codon. Results of EMSA analysis with 45-bp probes containing the predicted boxes are indicated under “Binding”; (+) and (−) indicate binding, or no binding, by MprA, respectively. Known MprA boxes refer to published sites confirmed by EMSAs, and located in the promoters for the given genes 25, 26, 28. d, p, i refer to distal, proximal, and intermediate promoter regions, respectively 26. Ratings of poor, moderate, and strong were obtained from published data 26, 28. “aux”, auxiliary site (see text for details).
(D–F) MprA was incubated with 45-bp probes containing predicted MprA boxes. (D) Probes containing Box A1 or Box A2 were incubated with the no MprA (Lanes 1 and 5), or 2.5 ug (Lanes 2 and 6), 5 ug (Lanes 3 and 7) or 7.5 ug (Lanes 4 and 8) of MprA. f, free probe; b, bound probe. (E) Comparison of MprA binding to Box E and a sigB promoter control. (−) and (+) symbols indicate incubation in the absence, or presence, of 7.5 ug MprA, respectively. (F) Binding of MprA to probes containing a native or mutagenized Box F. Lowercase italics in the sequences below the gel image indicate mutated bases. (−) and (+) symbols indicate incubation in the absence, or presence, of 2 ug MprA, respectively.