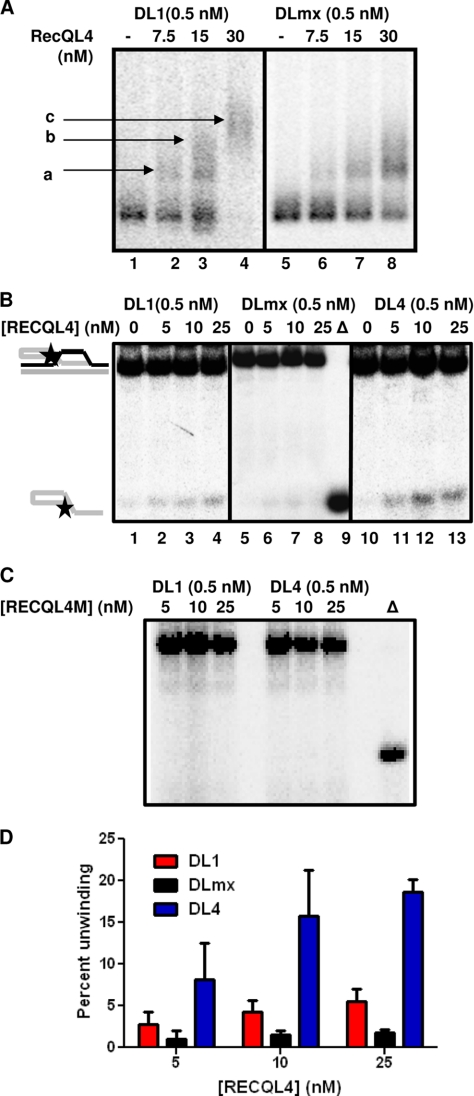
FIGURE 7.
Interaction of RECQL4 with telomeric and non-telomeric D-loops. A, autoradiogram showing binding of 0, 7.5, 15, and 30 nm RECQL4 with 0.5 nm DL1 (lanes 1–4) and 0.5 nm DLmx (lanes 5–8). Bound products are marked as a, b, and c. B, autoradiogram showing the unwinding activity of 0, 5, 10, and 25 nm RECQL4 on telomeric (lanes 1–4, 0.5 nm DL1) and non-telomeric D-loops (lanes 5–8, 0.5 nm DLmx) and oxidatively damaged telomeric D-loops (lanes 10–13, 0.5 nm DL4) in the presence of excess (25×) single-stranded DNA. Δ (lane 9), represents heat-denatured DLmx. C, autoradiogram showing the helicase activity of 5, 10, and 25 nm helicase-dead mutant RECQL4 (RECQL4M) on 0.5 nm DL1 and 0.5 nm DL4 in the presence of excess (25×) single-stranded DNA. D, histogram showing the unwinding activity of 0, 5, 10, and 25 nm RECQL4 on DL1, DLmx, and DL4 in the presence of excess (25×) single-stranded DNA. The error bars represent mean ± S.D., n = 3.