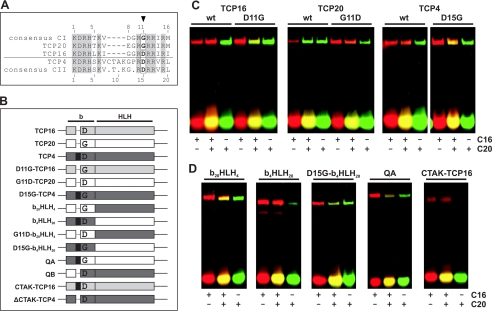
FIGURE 3.
Residue 11 of the class I TCP domain and residue 15 of the class II domain determine the binding preferences of TCP proteins. A, alignment of the basic regions of TCP16, TCP20, and TCP4, together with the consensus sequences of class I and class II TCP proteins. Conserved residues are shaded. Numbering indicates residue position within the respective TCP domain. The arrowhead indicates the residues that were exchanged in the mutant proteins under study. B, schematic structure of the mutants and chimeric proteins used in this study. Portions from TCP16, TCP20, or TCP4 are indicated in different colors. The black rectangle represents the 4-amino acid insertion of the TCP4 basic region. D or G indicates the presence of either Asp or Gly at position 11 (or 15 in proteins with the insertion). b and HLH indicate the basic region and the helix-loop-helix motif, respectively. The names of the proteins are indicated on the left. C, EMSA of TCP proteins and the respective mutants using C16, C20, or both oligonucleotides in the same binding reaction. C16 was 5′ end-labeled with Cy5, whereas C20 was 5′ end-labeled with 6-carboxyfluorescein. The images correspond to scans performed at the excitation and emission wavelengths of the corresponding fluorophores that were superimposed and colored in red and green for C16 and C20, respectively. D, EMSA similar to that in C with chimeric proteins with portions of TCP20 and TCP4. A TCP16 protein with the 4-amino acid insertion of TCP4 was also tested.