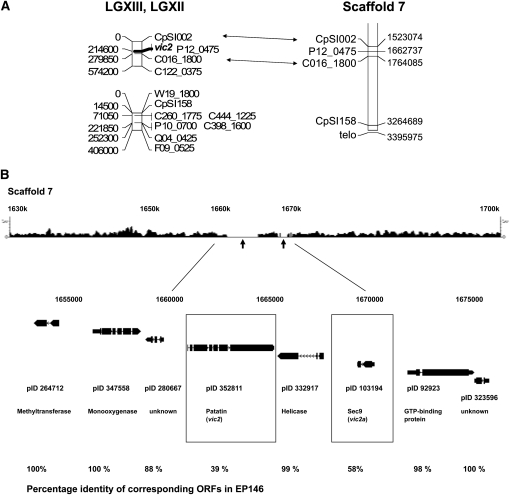
Figure 2 .
The candidate vic2 locus. (A) Alignment of LGXII and LGXIII of the C. parasitica genetic linkage map generated by Kubisiak and Milgroom (2006), the latter of which contains the position of the vic2 locus, with Scaffold 7 of version 2 of the C. parasitica genome sequence. The estimated physical distance (base pairs) of each marker from the end of the linkage group (shown on the left side of the linkage map) was calculated using the estimate of 1 cM = 14.5 kbp (Kubisiak and Milgroom 2006). Markers linking the physical and genetic maps are connected by double arrows. (B) The region of sequence polymorphism for C. parasitica strains EP155 and EP146 and machine-annotated ORFs located near the predicted vic2 locus. The top track shows the density plot of the 454 sequence reads (scale indicated at both ends) that match a corresponding portion of Scaffold 7 of the EP155 reference genome sequence assembly. The 10-kbp region of polymorphism between the EP155 and EP146 genome sequences in the region mapped for the vic2 locus is indicated by the absence of matching 454 sequence reads (arrows). The ORFs located within a 23-kbp region containing the polymorphic locus, with accompanying protein ID numbers, are shown below the 454 sequence read track. The candidate vic2 genes vic2-2 (patatin-like protein, pID 352811) and vic2a-2 (sec9-like protein, pID103194) are indicated by boxes. The percentages of amino acid identity for the corresponding EP155 and EP146 ORFs are indicated at the bottom.