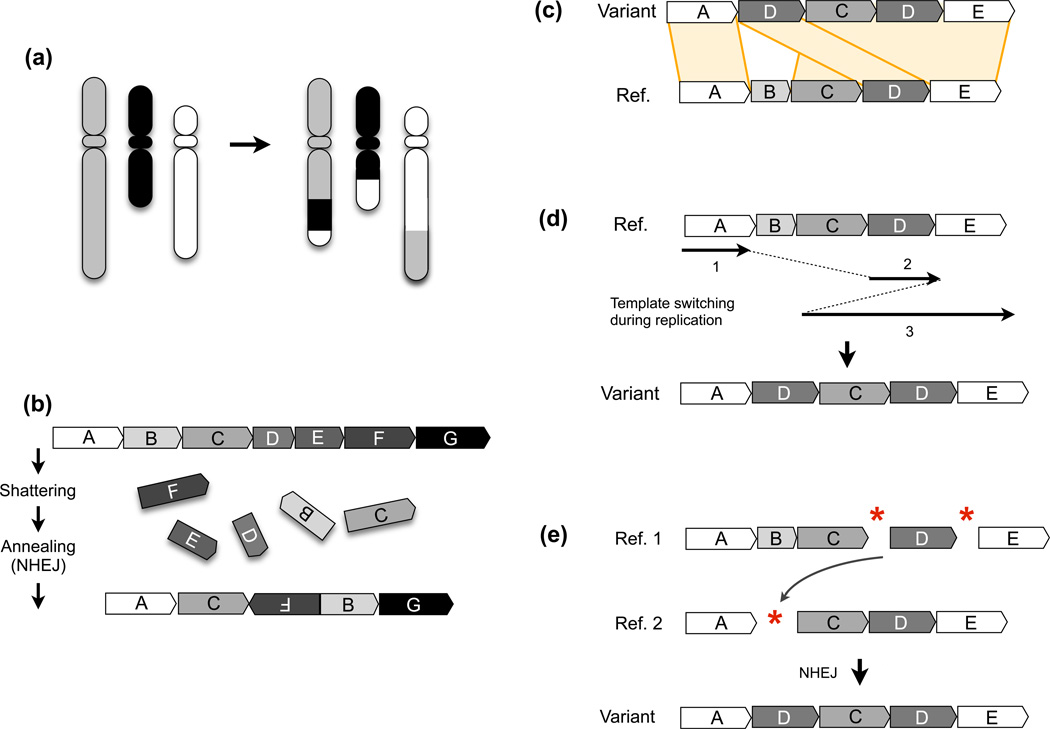
Figure 1.
High level view of complex rearrangements. (a) A depiction of a complex chromosomal rearrangement exhibiting 4 breakpoints from three differently-shaded chromosomes. (b) A complex rearrangement formed by chromothripsis, where a large section of a chromosome (segments B–F) is shattered and imprecisely stitched back together, leading to multiple intra-chromosomal rearrangements and loss of “D” and “E” segments. Chromosomal segments are indicated as blocks with different letters and shading, and the orientation by an arrow in each block. This diagram is loosely based on a figure in [94]. (c) A complex genomic rearrangement. The variant structure is shown at top and the reference genome (Ref.) below. Alignments are shown as orange blocks connecting the two sequences. This variant involves a deletion of “B” and a duplication of “D”, with “C” unaffected. (d) A model for how the variant structure in part (c) could arise through template switching during replication (e.g., FoSTeS/MMBIR), where continuously replicated segments are indicated as solid lines and template switches as dotted lines. (e) A model for how the variant structure in (c) could arise through NHEJ-mediated repair of three double strand DNA breaks. DNA breakages are indicated by red asterisks. In this example, a single DNA break destroys segment “B”, as can occur due to resection. The two copies of the reference locus (“Ref. 1” and “Ref. 2”) are present either as homologous chromosomes or sister chromatids.