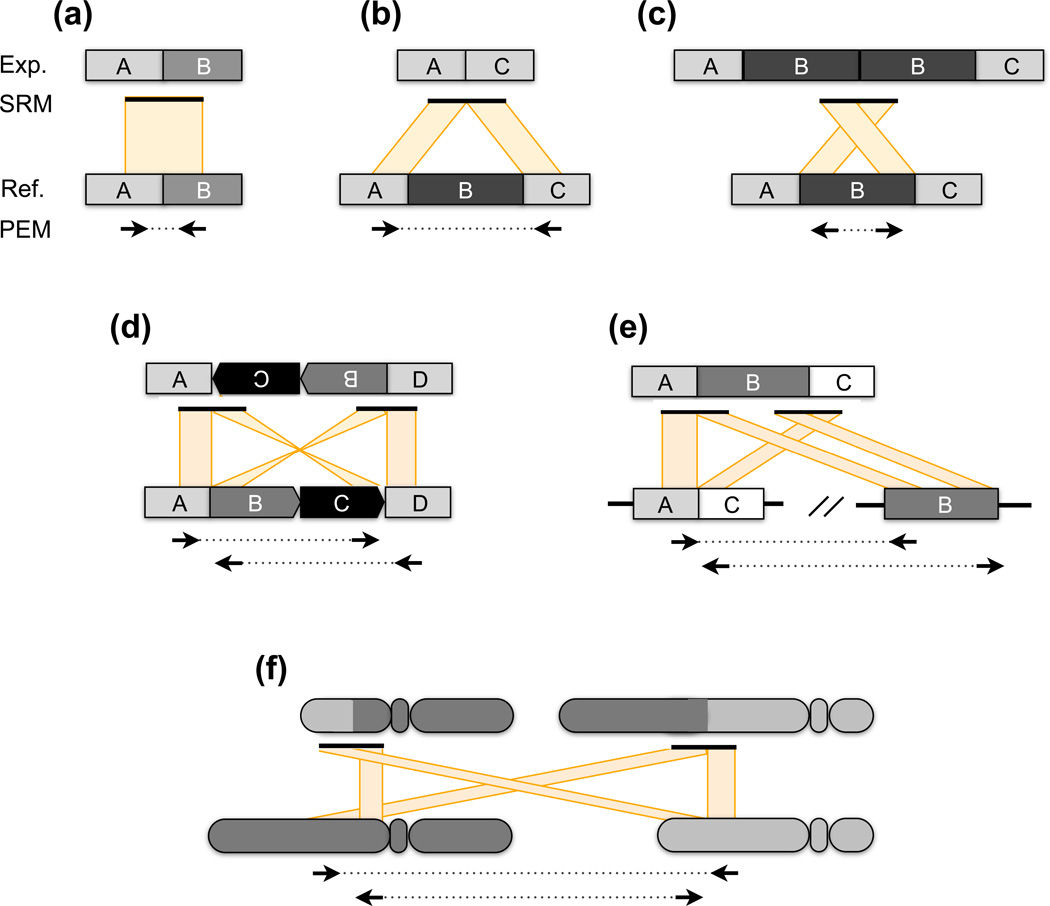
Figure 2.
Detecting canonical SV breakpoints through sequencing. When DNA sequences are collected from an experimental (Exp.) genome and aligned to a reference (Ref.) genome, each structural variant class generates a distinct alignment pattern. The patterns observed for paired-end mapping (PEM) and split-read mapping (SRM) are illustrated when both genomes have identical structure (a), and cases where the experimental genome contains a deletion (b), a tandem duplication (c), an inversion (d), a transposon insertion (e) and a reciprocal translocation (f). PEM relies upon readpairs whose unsequenced portion (dotted lines) spans a SV breakpoint. When aligned to the reference genome, the alignment distance and orientation of such readpairs indicates the type of rearrangement that has occurred. Reads that map to the plus strand are shown as right-facing arrows, those that map to the negative strand as leftward-facing arrows. All examples depict Illumina paired-end sequence data, where in the absence of SV the normal concordant orientation is plus for the leftmost read and minus for the rightmost read. Note that the expected orientation is different for Illumina mate-pair libraries and for other sequencing platforms such as SOLiD. In the case of a deletion (b), the readpairs ends will align much farther apart than expected for the DNA library. In contrast to PEM, SRM depends on contiguous sequences that contain an SV breakpoint. Consequently, the sequences before and after the breakpoint will align to disjoint regions of the reference genome. In contrast to PEM, breakpoints are identified at single-base resolution.