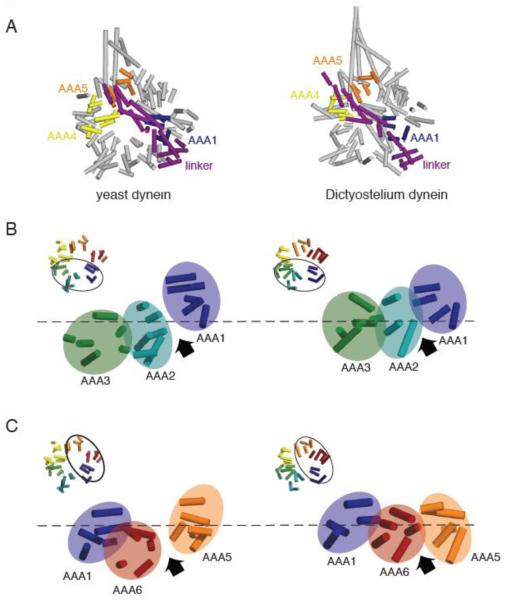
Figure 2.
(A) Comparison of the yeast (PDB ID: 3QMZ) and Dictyostelium dynein motor domain structures (PDB ID: 3AY1). The motor domains were superimposed by aligning the helices in the linker and AAA1, which superimpose well in the yeast and Dictyostelium structures. In the yeast dynein structure, the ring is more open and asymmetric than in the Dictyostelium dynein structure. Also, AAA5 is more shifted towards AAA4 in the yeast structure, resulting in a slight difference in the position where the N-terminus of the linker sits relative to the ring. (B) Comparison of yeast and Dictyostelium dynein AAA1-3. Asymmetry is more prominent and the AAA1-2 gap is wider in the yeast structure due to the different heights of AAA domains; specifically AAA2 is shifted further down with respect to AAA1. (C) Comparison of yeast and Dicytostelium dynein AAA1, 5, and 6. The AAA ring is more planar and the AAA5-6 gap smaller in the Dictyostelium structure. As discussed in the text, we speculate that the differences between yeast and Dictyostelium reflect distinct nucleotide occupancy in AAA1 (empty and ADP for yeast and Dictyostelium respectively). Note: the model for Dictyostelium dynein lacks several AAA helices that are present in the yeast model.