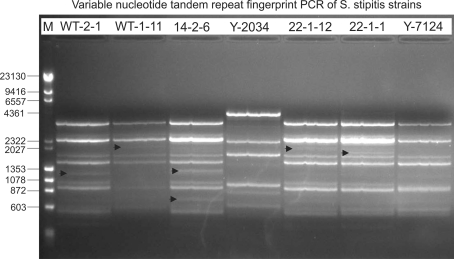
Fig. 3.
Variable nucleotide tandem repeat (VNTR) fingerprint of the changes to the genomic DNA of mutagenized S. (Pichia) stipitis strains WT-2-1, WT-1-11, 14-2-6, 22-1-12, and 22-1-1 compared to Saccharomyces cerevisiae NRRL Y-2034 and the wild-type (WT) strain, S. stipitis NRRL Y-7124. Resulting PCR products were obtained using the VNTR primer-generated fragments and are indicative of changes to the genomes of the mutagenized strains and analyzed on a 1% (w/v) agarose gel. Bionexus DNA base-pair markers (Lane M) are identified by length on the left of the figure. Bands present in mutagenized strains but not present in WT S. stipitis NRRL Y-7124 are indicated by arrows. Gel was run at 80 V on a Bio-Rad Power Pac 3000 system, and a high-resolution digital image file was generated with an AlphaImager 3400 gel imaging system using a trans-UV light (Alpha Innotech Corp, San Leandro, CA)