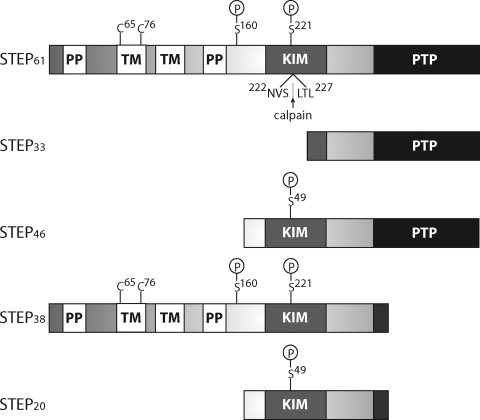
Fig. 2.
Schematic of STEP. There are four alternatively spliced variants of STEP (STEP61, STEP46, STEP38, and STEP20) and one calpain cleavage product (STEP33). STEP61 and STEP46 are the major isoforms expressed in the CNS. The KIM domain is required for substrate binding, and the consensus PTP sequence, [I/V]HCxAGxxR[S/T]G, is necessary for phosphatase activity. STEP38 and STEP20 do not contain the PTP sequence and are inactive variants of STEP with unknown function. It is possible that these two inactive isoforms function as dominant-negative variants that compete with active STEP variants for substrate binding, or they possess other functions yet to be discovered. STEP38 and STEP20 contain a unique 10-amino acid sequence at their carboxyl termini that is introduced during splicing. STEP33 is generated by calpain cleavage within the KIM domain between Ser224 and Leu225. Cleavage at this site disrupts the ability of STEP33 to interact with its substrates. STEP61 also contains an additional 172 amino acids in its amino-terminal region, which contains two transmembrane (TM) domains, two PP-rich regions, and an adjacent PEST sequence (not labeled). The TM regions target STEP61 to the endoplasmic reticulum, as well as the PSD. The KIM domain is required for binding to all substrates, whereas the PP regions impart substrate specificity. PKA phosphorylates STEP within the KIM domain (Ser221 and Ser49 on STEP61 and STEP46, respectively), as well as in the region adjacent to the PP regions (Ser160 on STEP61). Although the function of Ser160 phosphorylation on STEP61 remains unclear, current investigations are aimed at determining whether phosphorylation at this or other sites is a signal for calpain-mediated cleavage and/or ubiquitination. Finally, two cysteine residues Cys65 and Cys76 are found in the TM region that mediates STEP dimerization and reduces its phosphatase activity.