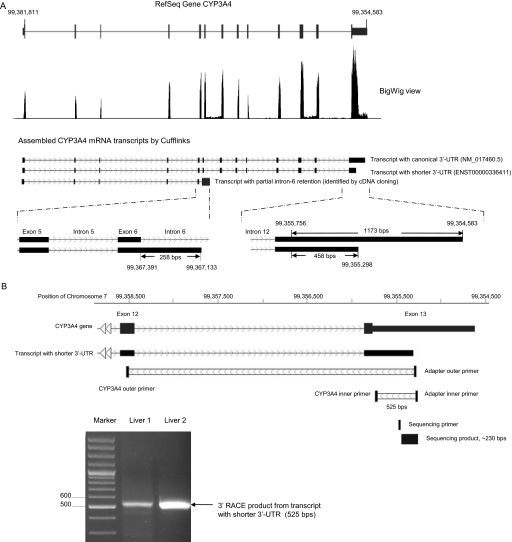
Fig. 2.
Identification of an alternative CYP3A4 mRNA transcript with a shorter 3′-UTR by RNA-Seq and validation by 3′RACE-PCR. A, expression of the CYP3A4 gene in HepaRG cells analyzed by RNA-Seq. Short-paired 100-bp RNA sequencing reads were aligned to the reference human genome hg19 by TopHat and viewed as distribution peaks by the UCSC genome browser. Cufflinks assembled the reads to the canonical and the shorter 3′-UTR transcripts and calculated their expression levels presented as FPKM. B, validation of the shorter 3′-UTR transcript by 3′RACE-PCR. First, total RNA of two liver tissues were reverse-transcribed with the poly(T)-containing adapter primer, subsequently followed by 3′RACE-PCR amplification with the adapter outer primer and the CYP3A4 outer primer and a nested PCR amplification with the adapter inner primer and the CYP3A4-specific inner primer. The PCR products were separated on a 2% agarose gel.