Fig. 2.
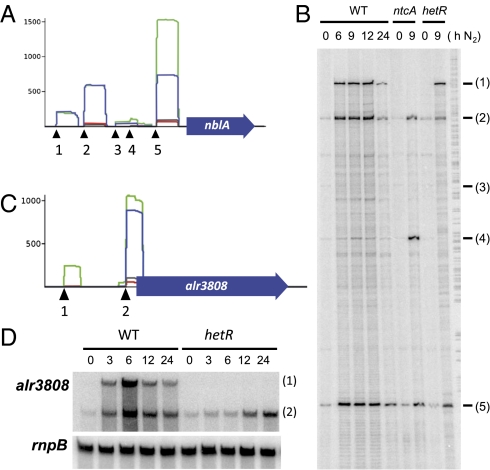
Analysis of genes with multiple TSS identified by dRNA-seq. RNA was isolated from ammonium-grown cells (lanes labeled 0) or from ammonium-grown cells incubated in the absence of combined nitrogen for the number of hours indicated. (A) Graphical representation of reads mapped to the promoter of nblA. The histograms correspond to the WT-0 (red), WT-8 (green), hetR-0 (black), and hetR-8 (blue) samples. (B) Primer extension analysis of the nblA mRNA in WT and mutant strains CSE2 (ntcA) and 216 (hetR). Samples contained 20 μg of RNA. The oligonucleotide used was complementary to positions +5 to −18 with respect to the translational start of nblA. The 5′ ends identified by dRNA-seq are numbered 1–5 (for positions, see Dataset 1, Table S1). (C) Graphical representation of reads mapped to the promoter of alr3808. (D) Northern blot analysis of the alr3808 mRNA in Anabaena 7120 and mutant strain 216 (hetR). Samples contained 10 μg of RNA. The probe used was an internal fragment of alr3808. rnpB (38) was used as a loading control.