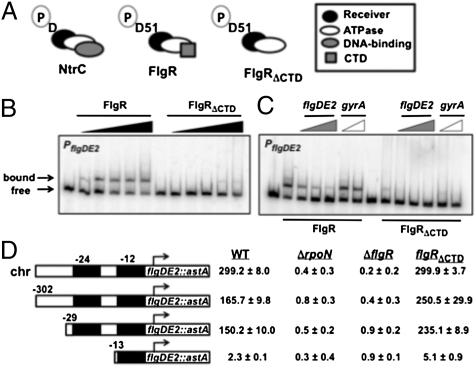
Fig. 1.
Analysis of the requirement of FlgR/DNA interactions for σ54-dependent flagellar gene expression. (A) Domain organization of E. coli NtrC, C. jejuni WT FlgR, and C. jejuni FlgRΔCTD. The phosphorylated aspartic acids (D or D51; P denotes phosphoryl group) in receiver domains are indicated. (B and C) EMSAs to analyze PflgDE2 DNA binding by FlgR proteins. (B) WT FlgR and FlgRΔCTD were used (from left to right) at 0, 0.1, 0.25, 0.5, 0.75, and 1 μM. (C) Increasing ratios of unlabeled PflgDE2 DNA (1:1, 5:1, or 10:1) or PgyrA DNA (1:1 or 10:1) to labeled PflgDE2 DNA were incubated with 1 μM of FlgR proteins. (D) Expression of flgDE2::astA with 5′ promoter truncations in WT C. jejuni or flgRΔCTD, ΔrpoN (Δσ54), or ΔflgR mutants. flgDE2::astA expression levels are reported as arylsulfatase units ± SD. All transcriptional fusions were on plasmids, except for the fusion at the native chromosomal flgDE2 locus (chr). The 5′ ends of fusions on plasmids are indicated relative to the transcriptional start site (arrow). Black boxes indicate the −24 and −12 σ54 binding sites essential for σ54-dependent gene expression.