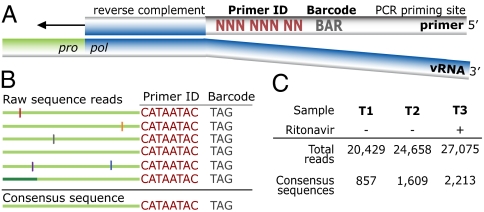
Fig. 1.
Tagging viral RNA templates with a Primer ID before PCR amplification and sequencing allows for direct removal of artifactual errors and identifies resampling. (A) A primer was designed to bind downstream of the protease coding domain. In the 5′ tail of the primer, a degenerate string of eight nucleotides created a Primer ID, allowing for 65,536 unique combinations. An a priori selected three nucleotide barcode was designed for the sample ID. Finally, a heterologous string of nucleotides with low affinity to the HIV-1 genome was included in the far 5′ end for use as the priming site in the PCR amplification. (B) PCR biases and sequencing error are introduced during amplification and sequencing of viral templates. Repetitive identification of the barcode and Primer ID allow for tracking of each templating event from a single tagged cDNA. As errors are minor components within the Primer ID population, forming a consensus sequence directly removes them, and corrects for PCR resampling. (C) HIV-1 RNA templates isolated from plasma samples from two pre- and one postintermittent ritonavir drug therapy were tagged, amplified, and deep sequenced. Tagged sequences containing full-length protease were used to create a population of consensus sequences when at least three sequences contained an identical barcode and Primer ID.