Abstract
We investigate yeast sex chromosome evolution by comparing genome sequences from 16 species in the family Saccharomycetaceae, including data from genera Tetrapisispora, Kazachstania, Naumovozyma, and Torulaspora. We show that although most yeast species contain a mating-type (MAT) locus and silent HML and HMR loci structurally analogous to those of Saccharomyces cerevisiae, their detailed organization is highly variable and indicates that the MAT locus is a deletion hotspot. Over evolutionary time, chromosomal genes located immediately beside MAT have continually been deleted, truncated, or transposed to other places in the genome in a process that is gradually shortening the distance between MAT and HML. Each time a gene beside MAT is removed by deletion or transposition, the next gene on the chromosome is brought into proximity with MAT and is in turn put at risk for removal. This process has also continually replaced the triplicated sequence regions, called Z and X, that allow HML and HMR to be used as templates for DNA repair at MAT during mating-type switching. We propose that the deletion and transposition events are caused by evolutionary accidents during mating-type switching, combined with natural selection to keep MAT and HML on the same chromosome. The rate of deletion accelerated greatly after whole-genome duplication, probably because genes were redundant and could be deleted without requiring transposition. We suggest that, despite its mutational cost, switching confers an evolutionary benefit by providing a way for an isolated germinating spore to reform spores if the environment is too poor.
The mating-type (MAT) locus is the only site in the Saccharomyces cerevisiae genome that is continually cleaved and repaired as part of the normal life cycle (1, 2). The MAT locus exists in two versions (idiomorphs) that contain either MATa or MATα genes, enabling it to specify three cell types: haploid a, haploid α, and diploid a/α. Mating-type switching is a programmed DNA rearrangement process that occurs in haploid cells and converts a MATa idiomorph into a MATα idiomorph, or vice versa. During switching, DNA at the MAT locus is removed and replaced with DNA copied from either the HML or HMR locus. HML and HMR are “silent cassettes” that store α-specific and a-specific sequence information, respectively, but are transcriptionally inactive due to chromatin modification (2–4).
The ability to switch mating type does not exist in all fungi, but originated independently at least twice (5): once in the family Saccharomycetaceae, which includes S. cerevisiae and Kluyveromyces lactis, and once in the Schizosaccharomycetaceae, which includes Schizosaccharomyces pombe. In the Saccharomycetaceae, switching evolved in a two-step process (6, 7). The first step was the origin of the HML and HMR cassettes, which occurred at the base of this family after it had diverged from other families such as Debaryomycetaceae and the Candida albicans clade (8). All species having silent cassettes, for example Lachancea waltii (9), are probably able to switch mating types using the homologous recombination machinery. However, in some clades, a second evolutionary step increased the rate and/or precision of switching by directing a dsDNA break to the MAT locus in cells that are about to switch. This second step occurred independently, by two different mechanisms, in two groups of yeast. In the “post-WGD” clade [species that underwent whole-genome duplication (10)] and their closest relatives such as Zygosaccharomyces rouxii, the dsDNA break is made by the HO endonuclease (6, 11). The HO gene does not exist outside this clade. In the genus Kluyveromyces, the dsDNA break is made by the excision of a mobile genetic element from the MATα idiomorph during the switch from MATα to MATa (12, 13). The mobile element contains a gene, α3, that is only present in Kluyveromyces. To switch in the opposite direction, from MATa to MATα, Kluyveromyces induces a dsDNA break at MATa by a different but uncharacterized mechanism (12).
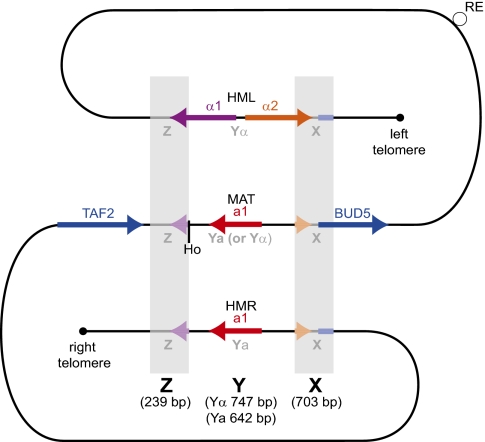
In all species that have silent cassettes, DNA repair at MAT is guided by two regions of sequence (the Z and X regions) that are almost identical between MAT, HML, and HMR. In S. cerevisiae, the Z region contains the 3′ end of the α1 gene, and the X region contains the 3′ end of α2 and the 5′ end of the neighboring chromosomal gene BUD5 (Fig. 1). The idiomorph-specific region between them is called Y, the two versions of which (Yα and Ya) have no sequence similarity to one another. In S. cerevisiae, switching begins when the HO endonuclease cleaves the Y–Z junction in the MAT locus (1, 2, 14). The old MAT-Y region is degraded. The Z and X sequences direct the use of HML or HMR as a template for repair, during which both strands of DNA at MAT-Y are newly synthesized in an error-prone fashion (15, 16). Repair is initiated by invasion of a 3′ end from MAT-Z into the HM donor (17, 18), so the first strand of the MAT-Y region is always synthesized in the direction from Z to X. Switching takes about 1 h (18).
Fig. 1.
Organization of the MAT, HML, and HMR loci on chromosome III of a MATa S. cerevisiae cell. The Z and X regions occur in three copies in parallel orientation and include parts of the α1, α2, and BUD5 genes. The Y region between them occurs in two versions (idiomorphs), Yα and Ya, which are completely dissimilar. This diagram is reversed relative to the standard S. cerevisiae orientation (2) to maintain compatibility with Figs. 2 and 3 despite species-specific inversions in S. cerevisiae (30). Note on nomenclature: We define X and Z as the regions that occur in three copies. In S. cerevisiae (2, 57), these are usually called X and Z1, and two duplicated regions that extend the similarity between MAT and HML (but not HMR) beyond them are called W and Z2. There are similar duplicated extensions at the outer edges of the triplicated regions in the other species studied here, but we did not see any consistent patterns of organization.
Switching does not occur during every cell cycle, but is a strategy that enables a “lonely” haploid yeast cell (that is, an isolated single cell that cannot find a partner of the opposite mating type) to produce diploid descendants (19–21). The haploid cell buds mitotically, the mother cell switches mating type, and the mother and daughter cells then mate to produce a homozygous diploid that can continue to replicate mitotically (22). In natural populations of Saccharomyces paradoxus, switching has been estimated to occur approximately once per 20,000 cell generations (23). The average generation time of natural yeast populations is not known (21), but generation times of 100 min (24), 100 h, or 100 d would correspond approximately to one mating-type switch per 4 y, 200 y, or 5,000 y, respectively. Even at the lowest of these rates, two yeast species that diverged 10 million y ago would each have gone through 2,000 switches since they shared a common ancestor, so switching needs to be efficient and accurate. For species that grow primarily as haploids, the rate of switching may be much higher. In this paper, we report evidence that switching errors do accumulate along evolutionary lineages and have had a profound effect on the structure of the MAT-containing chromosome in post-WGD species.
Results
Conservation of MAT–HML Linkage.
We compared MAT locus organization in 16 species of the family Saccharomycetaceae (25). We augmented existing data with genome sequences for seven species: two each from the post-WGD genera Kazachstania, Tetrapisispora, and Naumovozyma, and one from the non-WGD genus Torulaspora. The data support previous hypotheses that the three-cassette structure (MAT, HML, HMR) originated at the base of the Saccharomycetaceae (6–8), the HO endonuclease is younger than the three-cassette structure (6, 12), and the loss of the MATa2 gene (6, 26, 27) occurred on the same branch of the phylogenetic tree as the WGD. No losses of the MATa1, MATα1, or MATα2 genes occur in the Saccharomycetaceae species, in contrast to the multiple losses of MAT genes in the Candida clade (28, 29).
Among the 14 species in which mating-type switching appears to be possible, we find that MAT and HML are always on the same chromosome (86–310 kb apart) and the genotype of HML is always α. HMR is often on a different chromosome (30), and some species have two HMR loci (31). HML and HMR are usually but not invariably subtelomeric. The conservation of HML and MAT in cis, and of the α genotype at HML, is probably due to conservation of the recombination enhancer (RE) site among species. The RE, which has so far only been found in S. cerevisiae (32, 33), is located in the interval between HML and MAT. It increases the frequency of productive switching by biasing the choice of donor (32), and operates by binding the α2 protein (34, 35). The two species that may be unable to switch mating type are L. kluyveri, which has no HML or HMR (6, 36), and Kazachstania africana, which appears to have separate MATα and MATa loci due to a genomic rearrangement and has lost HML, HMR, and the HO endonuclease gene.
Turnover of Z and X Regions.
Although the MAT loci of most of the species are organized in a manner analogous to that of S. cerevisiae, the detailed structure of the Z and X regions varies extensively in terms of which MAT genes and neighboring chromosomal genes extend into them (Fig. 2). The X regions of S. cerevisiae and Kazachstania naganishii, for instance, have nothing in common. This variation is surprising, because the Z and X regions are virtually identical among the three copies within each genome, and were previously found to be among the most slowly evolving sequences in the genome (with >96% identity) among four species in the genus Saccharomyces that are separated by tens of millions of years (37). Therefore, the Z- and X-region sequences have low rates of nucleotide substitution but can be completely replaced. There is an evolutionary requirement for triplicated sequences flanking MAT, HML, and HMR to guide mating-type switching, but the requirement is for triplication per se and not for any particular sequence.
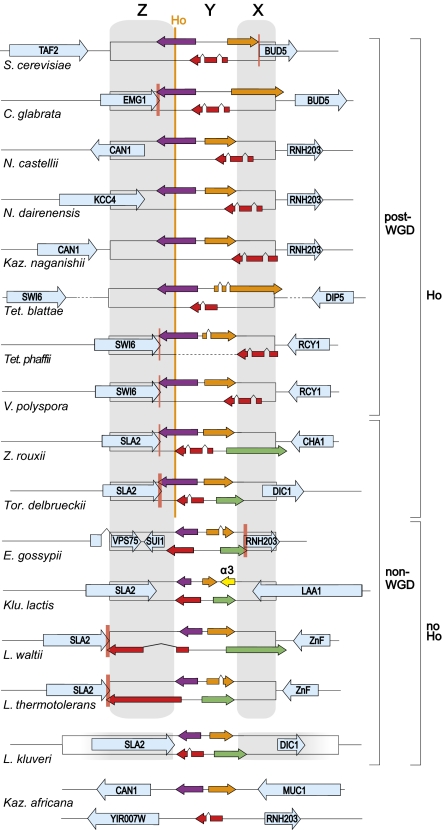
Fig. 2.
Schematic organization (not to scale) of the MAT locus in 16 species. Both possible versions of the Y region are shown for each species. Yα contains the genes α1 (purple) and α2 (orange). Ya contains the genes a1 (red) and a2 [green, only in non-WGD species (6, 26)]. Caret symbols indicate introns. Gray shading indicates the extent of the Z and X regions. HO endonuclease, where present, cleaves the MAT locus at the Y–Z boundary at a site in the α1 gene. Flanking chromosomal genes are shown in blue. Pink vertical bars indicate gene overlaps (broad bars) or intergenic distances ≤5 bp (narrow bars). In L. kluyveri there are no HML and HMR cassettes (36), but the sequenced strain is diploid so only the inner boundaries of Z and X are defined. In Kazachstania africana there are two MAT-like regions and no HO gene. The dashed line for Tetrapisispora phaffii Ya represents zero length of sequence. Fig. S3 shows the same regions drawn to scale.
A general principle of MAT locus organization apparent from Fig. 2 is that the idiomorph-specific region Yα must contain parts of both the α1 and α2 genes, and Ya must contain parts of the a1 and (where present) a2 genes, so that the gene fragments in the MAT-Z and MAT-X regions are incapable of expression in cells with the “wrong” genotype. Beyond this principle, however, it does not seem to matter which MAT genes extend into Z and X (Fig. 2), although in species with the HO endonuclease the Y–Z junction has been stabilized to a site in MATα1. Tetrapisispora phaffii is puzzling because it seems to violate the principle: It has no Ya region (there is no DNA between the Z and X regions in its MATa idiomorph), so it is not clear how (or whether) MATa1 expression is prevented in MATα cells of this species.
Collision and Truncation of Chromosomal Genes Flanking MAT.
The Z and X regions often include parts of flanking chromosomal genes whose functions are not related to cell identity (colored blue in Fig. 2), again with much variation among species. These genes are partially duplicated at HML and HMR. Remarkably, there is often almost no intergenic DNA between the flanking genes and the MAT genes, and in some cases they overlap (Fig. 2 and Fig. S1). Some flanking genes are truncated, such as S. cerevisiae BUD5, whose start codon overlaps the stop codon of MATα2. The Bud5 protein is only half the length of its orthologs in other species, lacking an SH3 domain at its N terminus (38). SLA2, SWI6, and LAA1 in other species are all similarly truncated at their ends closest to MAT (Fig. S2). These features are all suggestive of a process that tends to delete nonessential DNA beside the MAT locus.
Progressive DNA Deletion Beside MAT.
To investigate how the MAT locus acquired different flanking genes in different post-WGD species, we compared the genomes to the “Ancestral” gene order (39) inferred to have existed just before WGD occurred. In the Ancestral genome nomenclature (39), HML and MAT are on chromosome 1 (Anc_1), with HMLα1 and HMLα2 being the first two genes on this chromosome (Anc_1.1 and Anc_1.2) and the MAT locus about 120 genes farther along (positions Anc_1.120 to Anc_1.122) (Fig. 3). The genes ancestrally flanking MAT are SLA2 and DIC1, an arrangement that appears to be quite old and stable because it is conserved in Komagataella phaffii (Pichia pastoris) (40) and Ogataea (Hansenula) polymorpha (6). Ancestral chromosome 1 was duplicated as part of WGD, giving rise to two daughter chromosomes. We call one daughter the “MAT chromosome” because it retained the MAT and HML loci, and the other the “non-MAT chromosome” because it lost its copies of these loci. Both chromosomes underwent further rearrangement after WGD, but in each post-WGD species the chromosomal regions derived from the MAT and non-MAT chromosomes can be identified by tracing the products of each rearrangement event (39), and are shown in Fig. 3.
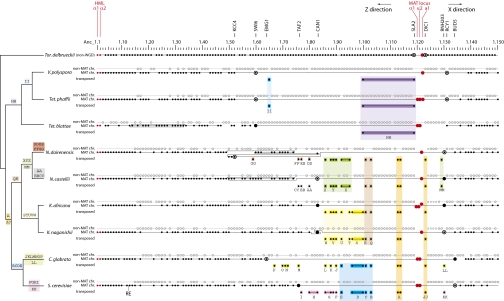
Fig. 3.
Progressive loss of genes flanking the MAT locus by deletion and transposition. The scale indicates gene positions along part of Ancestral chromosome 1, from Anc_1.1 to Anc_1.150. Each circle represents a gene, with HML and MAT genes in red (each genome sequence is arbitrarily either MATα or MATa). Horizontal lines connect genes that are currently neighbors; zigzags show inversions. For each post-WGD species, genes are assigned to three groups: those derived from the MAT chromosome (the chromosome that retained the MAT locus after WGD; black circles); those derived from the non-MAT chromosome (the paralogous chromosome that lost the MAT locus after WGD; open circles); and those that transposed from the MAT chromosome to other places in the genome (letters A–Z and AA–MM; colored backgrounds). Each transposition can be inferred to have occurred on a particular branch of the phylogenetic tree on the left, based on the clade of species that share the insertion site, as shown by the different colors. Genes named above the scale are the current neighbors of the MAT locus in the species shown here; these genes are identified by bullseyes (for flanking genes that extend into the Z or X regions) or large black circles. Due to a large inversion in S. cerevisiae that spans the MAT locus and the centromere (30), the Z and X directions as indicated at the top correspond to rightward and leftward, respectively, on chromosome III. More details are given in Fig. S5.
Strikingly, large deletions are seen on the MAT chromosome in each post-WGD species, beginning at the MAT locus and extending in the Z direction (leftward as drawn in Fig. 3). These deletions brought genes that were originally farther away in the interval between HML and MAT into direct proximity with MAT. In Vanderwaltozyma polyspora, for example, SWI6 (Anc_1.60) is now the neighbor of MATa1 (Anc_1.122) on the MAT chromosome, and almost all of the Ancestral genes between them were retained on the non-MAT chromosome instead (Fig. 3). This nonrandom distribution of genes between sister chromosomal regions contrasts with the usual pattern of gene losses after WGD (31, 41). The most obvious explanation is that 60 consecutive genes were removed from the MAT chromosome in the V. polyspora lineage by a deletion(s) that occurred soon after WGD, at a time when most of its genome was still duplicated. The deletions have different endpoints in different post-WGD species, so that among the nine post-WGD species in Fig. 3 the current neighbors of MAT on the Z side are KCC4 (Anc_1.52), SWI6 (Anc_1.60), EMG1 (Anc_1.64), TAF2 (Anc_1.76), and CAN1 (Anc_1.83). A similar but less extensive deletion process has occurred on the other (X) side, where the genes flanking MAT are RNH203 (Anc_1.130), RCY1 (Anc_1.131), and BUD5 (Anc_1.134) in different post-WGD species. In Tetrapisispora blattae, a translocation has joined the X side of MAT to a telomeric region. Rearrangements like this probably cannot occur on the Z side due to the evolutionary constraint to maintain MAT and HML on the same chromosome.
In contrast to the situation for post-WGD species, none of the non-WGD species show large deletions beside the MAT locus. They all retain an organization similar to Torulaspora delbrueckii, which is shown for illustration in Fig. 3. In different non-WGD species, the genes neighboring MAT on the Z side are SLA2 (Anc_1.119) and SUI1 (Anc_1.118), and on the X side DIC1 (Anc_1.123), LAA1 (Anc_1.127), RNH203 (Anc_1.130), and an unnamed zinc-finger gene located between Anc_1.123 and 1.124 (Fig. 2). In Z. rouxii, similarly to Tetrapisispora blattae, a translocation has joined the X side of MAT to a telomeric region containing CHA1.
Gene Transpositions Provide a Timeline.
Instead of being deleted, some genes transposed away from the vicinity of the MAT locus. For instance, S. cerevisiae JJJ3 (Anc_1.113) is not found in the expected region of the MAT or non-MAT chromosome (parts of chromosomes III and XIV, respectively), but instead is on chromosome X (YJR097W). JJJ3 and its neighbor YJR098C (Anc_1.114) transposed from the MAT chromosome to a new genomic location descended from Ancestral chromosome 7, where they were inserted between genes Anc_7.468 (YJR096W) and Anc_7.470 (YJR099W). We found 39 separate such events of transposition away from MAT and use letters A–Z and AA–MM to identify them (Fig. 3). Each transposition event moved one to three genes. Of the 39 events, 35 are on the Z side of MAT and 4 are on the X side.
The transposition of JJJ3 and YJR098C to the site on Ancestral chromosome 7 (event A in Fig. 3) is shared by the genomes of six post-WGD species, so it must have occurred in their common ancestor. Further to the left (Z side) of the MAT locus, events B, C, D, and E are transpositions shared by S. cerevisiae and C. glabrata (they have the same four insertion sites), but not other species. Further left again, events F, G, H, and I are unique to S. cerevisiae, and then we reach the gene (TAF2, Anc_1.76) that is the current neighbor of MAT in S. cerevisiae. A similar pattern is seen in each other post-WGD species (Fig. 3 and Table S1). It is evident that the genes transposed in a particular order, with those closest to Anc_1.120 moving before those further to the left, over a long time period during which the post-WGD lineages diverged from one another, as shown by the phylogenetic tree in Fig. 3.
We therefore infer that the MAT locus tends to cause the deletion or transposition of the gene that is its immediate neighbor on the Z side. When one neighbor is removed, the next comes under attack. During the 100–200 million y since WGD, this process has removed a series of 44–60 MAT-neighboring genes in different post-WGD species. On the X (right) side, only four transpositions are seen, but again an older transposition (event JJ) involved a gene that was ancestrally closer to the MAT locus than the younger transpositions (events KK–MM).
Discussion
We hypothesize that the evolutionary deletions, gene truncations, and transpositions beside the MAT locus were made during recovery from occasional accidents that occurred during mating-type switching. DNA synthesis during switching in S. cerevisiae is highly prone to errors, including microhomology-mediated jumps to ectopic templates (16). The evolutionary deletions resemble the long one-sided deletions found extending up to 12 kb from the HO site, in the Z direction, in about 2% of S. cerevisiae cells in experiments by Yu and Gabriel (42) in which the cleaved chromosome was repaired by microhomology-mediated end joining (MMEJ) because no donor sequence was available. During switching in S. cerevisiae, the HO double-strand break is processed (resected) to generate a long single-stranded tail that can include all of the Z region and extend into the flanking gene (TAF2) beyond it (17). If this tail broke and lost the Z region, no homologous donor would be available; to repair the chromosome in a way that satisfies the constraint (imposed by the RE) to keep MAT and HML in cis would require religation by MMEJ, deleting part or all of TAF2. If instead the tail invaded some other place in the genome, it could cause transposition of TAF2 before the HML–MAT linkage is restored. The greater extent of deletions and transpositions seen on the Z side than on the X side (Fig. 3) may be because DNA-strand exchange initiates in the Z region (17, 18). Successful repair of the chromosome would also require the new sequence flanking MAT to be copied to HML and HMR to become a new Z region; the fact that different chromosomal genes are incorporated into the Z and X regions in different species (Fig. 2) shows that such a feedback mechanism exists.
We infer that a tendency to delete DNA beside the MAT locus exists in non-WGD species as well as post-WGD species, because we see flanking gene truncations and some small gene deletions in non-WGD species (Fig. 2 and Fig. S2) (43). However, the effects of the deletion process are much more drastic in post-WGD species (Fig. 3). We hypothesize that the difference is because WGD brought redundancy into the genome. Suddenly no genes beside the MAT locus were essential because they all had a second copy on the non-MAT chromosome, so large deletions were possible. As time progressed, duplicated genes were lost from throughout the post-WGD genome, and some genes in the interval between HML and MAT became single-copy. We propose that when the deletion process brought MAT adjacent to an essential single-copy gene, the process stalled until the gene transposed away from beside MAT. It is notable that some genes such as TAM41 (Anc_1.86) transposed independently in multiple lineages to different genomic sites (events G, L, W, and Z; Fig. 3 and Table S1). We suggest that its paralog on the non-MAT chromosome was lost soon after WGD, making TAM41 essential and so requiring it to be relocated in each lineage when MAT encroached on it. Some patterns of transposition (events HH, AA, BB, and CC) also indicate that a gene can be “trapped” in the Z region for a period while genes further to its left are deleted. Eremothecium gossypii SUI1 (Anc_1.118) may be an example of a trapped gene because CWC25 (Anc_1.117) has transposed from between it and VPS75 (Anc_1.116) (Fig. 2).
Our analysis suggests that errors during mating-type switching, combined with natural selection to keep MAT and HML on the same chromosome, have subjected the genes flanking the MAT locus to a continual process of attempted deletion and occasional transposition during evolution. Deletions were rampant in the immediate aftermath of WGD, but the rate at which MAT is moving toward HML is slowing (Fig. S4) because more genes are single-copy and need to be rescued by transposition. The deletion process removes genes and is therefore likely to impact on the biology of the species in which it occurs. One likely gene loss due to this process was a cyclin gene similar to C. albicans CCN1 (44), which has no ortholog in S. cerevisiae. This gene is located between positions Anc_1.77 and Anc_1.78 in non-WGD species. It has been lost from all post-WGD genomes, except in the genus Kazachstania, where it survives because the MAT locus has only deleted Z-ward as far as Anc_1.83 in that genus (Fig. 3). Another possible casualty is the MATa2 gene itself, whose loss led to rewiring of the cell identity pathway (26, 27).
Sex chromosomes are subject to unique evolutionary processes and mechanisms (5, 45–47). Our observations about the yeast MAT chromosome are reminiscent of the movement of genes out of the mammalian X chromosome (48, 49), but unlike that process we do not suggest that the “out-of-MAT” gene movements are driven by natural selection. Instead, we propose a mechanical explanation: that mating-type switching is accident-prone, and that recovery from these accidents erodes the flanking chromosomal DNA. The fact that switching has been an evolutionarily successful strategy (23) implies that it must confer a benefit that outweighs the mutational costs of the deletions described here and of the error-prone DNA synthesis that occurs during switching (16). What is this benefit? Unlike recombination, switching does not create or maintain any genetic diversity. And because switching occurs both in species that grow primarily as diploids (such as S. cerevisiae and most post-WGD lineages) and in others that grow primarily as haploids and sporulate immediately after mating (such as Kluyveromyces lactis and most non-WGD lineages), the benefit cannot simply be one of diploidy over haploidy. We suggest that the benefit of switching may be that, in effect, it makes spore germination reversible. Consider a single isolated spore that finds itself in a poor environment. In a yeast species that cannot switch mating types, if the spore germinates it commits itself irreversibly (50) to mitotic growth until it finds a mating partner. If the environment is too harsh, this cell lineage will go extinct. In contrast, in a species that can switch, an isolated spore that germinates in a harsh environment can form new spores genetically identical to itself after just two mitotic cell divisions (51), followed by switching, mating, and sporulation. In this way, mating-type switching may have the benefit of allowing spores to test environments of uncertain quality. In poor environments one could envisage spores going through repeated cycles of germination, switching, and resporulation, possibly leading to periodic bursts of switching and increased rates of DNA erosion at the MAT locus.
Materials and Methods
Sequencing.
The genomes were sequenced using Roche FLX technology with the aim of achieving high contiguity and establishing the order of genes along chromosomes. We sequenced the type strains, purchased from the Centraalbureau voor Schimmelcultures (CBS), of these species in the family Saccharomycetaceae (25): Tetrapisispora phaffii (CBS 4417; 17 scaffolds), Tetrapisispora blattae (CBS 6284; 10 scaffolds), N. dairenensis (CBS 421; 12 scaffolds), Kazachstania africana (CBS 2517; 12 scaffolds), Kazachstania naganishii (CBS 8797; 13 scaffolds), and Torulaspora delbrueckii (CBS 1146; 7 scaffolds). We also completed the sequence of N. castellii (CBS 4309; previously called S. castellii or Naumovia castellii; 10 scaffolds), which was draft-sequenced by Cliften et al. (52, 53). Sequencing was done under contract by Eurofins MWG Operon. Each genome was sequenced to >20× coverage (>1 million reads) using a Roche GS FLX instrument with titanium reagents, with a mixture of paired (3-kb, 8-kb, and 20-kb genomic DNA inserts; 1/4 of data each) and unpaired (1/4 of data) sequence reads. Data were assembled into contigs and scaffolds using the Celera assembler (54). All intercontig joins in the scaffold data were checked manually by reference to the paired-end reads and by comparison with other species. All scaffolds appear to correspond to complete chromosomes, except for one unplaced 15-kb scaffold in Tetrapisispora phaffii. Ribosomal DNA was assembled and integrated into the scaffolds manually. Mitochondrial genomes were not assembled.
Annotation.
We developed a pipeline, to be described in detail elsewhere, that uses gene order and sequence data from the Yeast Gene Order Browser (YGOB) database (55) to annotate yeast genomes. The pipeline uses an approach based on TBLASTN (56) to overcome frameshift sequencing errors.
Data Access.
Genomes can be viewed in the YGOB database (http://wolfe.gen.tcd.ie/ygob).
Supplementary Material
Acknowledgments
We thank A. Rourke and F. S. Dietrich for help, and G. Butler and two referees for constructive comments. This work was supported by Science Foundation Ireland and the European Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the European Molecular Biology Laboratory Nucleotide Sequence Database (accession nos. HE576752–HE576761 and HE580267–HE580278).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112808108/-/DCSupplemental.
References
- 1.Strathern JN, et al. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell. 1982;31(1):183–192. doi: 10.1016/0092-8674(82)90418-4. [DOI] [PubMed] [Google Scholar]
- 2.Haber JE. Mating-type gene switching in Saccharomyces cerevisiae. Annu Rev Genet. 1998;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- 3.Hicks JB, Strathern JN, Herskowitz I. In: DNA Insertion Elements, Plasmids and Episomes. Bukhari A, Shapiro J, Adhya S, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1977. pp. 457–462. [Google Scholar]
- 4.Herskowitz I, Rine J, Strathern JN. In: The Molecular and Cellular Biology of the Yeast Saccharomyces. Jones EW, Pringle JR, Broach JR, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1992. pp. 583–656. [Google Scholar]
- 5.Lee SC, Ni M, Li W, Shertz C, Heitman J. The evolution of sex: A perspective from the fungal kingdom. Microbiol Mol Biol Rev. 2010;74:298–340. doi: 10.1128/MMBR.00005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler G, et al. Evolution of the MAT locus and its Ho endonuclease in yeast species. Proc Natl Acad Sci USA. 2004;101:1632–1637. doi: 10.1073/pnas.0304170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett RJ, Johnson AD. Mating in Candida albicans and the search for a sexual cycle. Annu Rev Microbiol. 2005;59:233–255. doi: 10.1146/annurev.micro.59.030804.121310. [DOI] [PubMed] [Google Scholar]
- 8.Dujon B, et al. Genome evolution in yeasts. Nature. 2004;430(6995):35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 9.Di Rienzi SC, et al. Genetic, genomic, and molecular tools for studying the protoploid yeast, L. waltii. Yeast. 2011;28(2):137–151. doi: 10.1002/yea.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 11.Kostriken R, Heffron F. The product of the HO gene is a nuclease: Purification and characterization of the enzyme. Cold Spring Harb Symp Quant Biol. 1984;49:89–96. doi: 10.1101/sqb.1984.049.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Barsoum E, Martinez P, Aström SU. α3, a transposable element that promotes host sexual reproduction. Genes Dev. 2010;24(1):33–44. doi: 10.1101/gad.557310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rusche LN, Rine J. Switching the mechanism of mating type switching: A domesticated transposase supplants a domesticated homing endonuclease. Genes Dev. 2010;24(1):10–14. doi: 10.1101/gad.1886310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haber JE. Transpositions and translocations induced by site-specific double-strand breaks in budding yeast. DNA Repair (Amst) 2006;5:998–1009. doi: 10.1016/j.dnarep.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 15.Ira G, Satory D, Haber JE. Conservative inheritance of newly synthesized DNA in double-strand break-induced gene conversion. Mol Cell Biol. 2006;26:9424–9429. doi: 10.1128/MCB.01654-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329(5987):82–85. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White CI, Haber JE. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9:663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hicks WM, Yamaguchi M, Haber JE. Real-time analysis of double-strand DNA break repair by homologous recombination. Proc Natl Acad Sci USA. 2011;108:3108–3115. doi: 10.1073/pnas.1019660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herskowitz I. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol Rev. 1988;52:536–553. doi: 10.1128/mr.52.4.536-553.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Replansky T, Koufopanou V, Greig D, Bell G. Saccharomyces sensu stricto as a model system for evolution and ecology. Trends Ecol Evol. 2008;23:494–501. doi: 10.1016/j.tree.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Greig D, Leu JY. Natural history of budding yeast. Curr Biol. 2009;19:R886–R890. doi: 10.1016/j.cub.2009.07.037. [DOI] [PubMed] [Google Scholar]
- 22.Mortimer RK. Evolution and variation of the yeast (Saccharomyces) genome. Genome Res. 2000;10:403–409. doi: 10.1101/gr.10.4.403. [DOI] [PubMed] [Google Scholar]
- 23.Tsai IJ, Bensasson D, Burt A, Koufopanou V. Population genomics of the wild yeast Saccharomyces paradoxus: Quantifying the life cycle. Proc Natl Acad Sci USA. 2008;105:4957–4962. doi: 10.1073/pnas.0707314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 25.Kurtzman CP. In: The Yeasts: A Taxonomic Study. 5th Ed. Kurtzman CP, Fell JW, Boekhout T, editors. Vol 2. Amsterdam: Elsevier; 2011. pp. 293–307. [Google Scholar]
- 26.Tsong AE, Miller MG, Raisner RM, Johnson AD. Evolution of a combinatorial transcriptional circuit: A case study in yeasts. Cell. 2003;115:389–399. doi: 10.1016/s0092-8674(03)00885-7. [DOI] [PubMed] [Google Scholar]
- 27.Tsong AE, Tuch BB, Li H, Johnson AD. Evolution of alternative transcriptional circuits with identical logic. Nature. 2006;443:415–420. doi: 10.1038/nature05099. [DOI] [PubMed] [Google Scholar]
- 28.Butler G, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reedy JL, Floyd AM, Heitman J. Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex. Curr Biol. 2009;19:891–899. doi: 10.1016/j.cub.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fabre E, et al. Comparative genomics in hemiascomycete yeasts: Evolution of sex, silencing, and subtelomeres. Mol Biol Evol. 2005;22:856–873. doi: 10.1093/molbev/msi070. [DOI] [PubMed] [Google Scholar]
- 31.Scannell DR, et al. Independent sorting-out of thousands of duplicated gene pairs in two yeast species descended from a whole-genome duplication. Proc Natl Acad Sci USA. 2007;104:8397–8402. doi: 10.1073/pnas.0608218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X, Haber JE. A 700 bp cis-acting region controls mating-type dependent recombination along the entire left arm of yeast chromosome III. Cell. 1996;87:277–285. doi: 10.1016/s0092-8674(00)81345-8. [DOI] [PubMed] [Google Scholar]
- 33.Zhou Z, Sun K, Lipstein EA, Haber JE. A Saccharomyces servazzii clone homologous to Saccharomyces cerevisiae chromosome III spanning KAR4, ARS 304 and SPB1 lacks the recombination enhancer but contains an unknown ORF. Yeast. 2001;18:789–795. doi: 10.1002/yea.724. [DOI] [PubMed] [Google Scholar]
- 34.Szeto L, Fafalios MK, Zhong H, Vershon AK, Broach JR. α2p controls donor preference during mating type interconversion in yeast by inactivating a recombinational enhancer of chromosome III. Genes Dev. 1997;11:1899–1911. doi: 10.1101/gad.11.15.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu C, et al. Mcm1 regulates donor preference controlled by the recombination enhancer in Saccharomyces mating-type switching. Genes Dev. 1998;12:1726–1737. doi: 10.1101/gad.12.11.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Souciet JL, et al. Génolevures Consortium Comparative genomics of protoploid Saccharomycetaceae. Genome Res. 19:1696–1709. doi: 10.1101/gr.091546.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 38.Wong S, Fares MA, Zimmermann W, Butler G, Wolfe KH. Evidence from comparative genomics for a complete sexual cycle in the ‘asexual’ pathogenic yeast Candida glabrata. Genome Biol. 2003;4(2):R10. doi: 10.1186/gb-2003-4-2-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gordon JL, Byrne KP, Wolfe KH. Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome. PLoS Genet. 2009;5:e1000485. doi: 10.1371/journal.pgen.1000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Schutter K, et al. Genome sequence of the recombinant protein production host Pichia pastoris. Nat Biotechnol. 2009;27:561–566. doi: 10.1038/nbt.1544. [DOI] [PubMed] [Google Scholar]
- 41.Dietrich FS, et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- 42.Yu X, Gabriel A. Ku-dependent and Ku-independent end-joining pathways lead to chromosomal rearrangements during double-strand break repair in Saccharomyces cerevisiae. Genetics. 2003;163:843–856. doi: 10.1093/genetics/163.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wendland J, Walther A. Ashbya gossypii: A model for fungal developmental biology. Nat Rev Microbiol. 2005;3:421–429. doi: 10.1038/nrmicro1148. [DOI] [PubMed] [Google Scholar]
- 44.Whiteway M, Dignard D, Thomas DY. Dominant negative selection of heterologous genes: Isolation of Candida albicans genes that interfere with Saccharomyces cerevisiae mating factor-induced cell cycle arrest. Proc Natl Acad Sci USA. 1992;89:9410–9414. doi: 10.1073/pnas.89.20.9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bachtrog D. A dynamic view of sex chromosome evolution. Curr Opin Genet Dev. 2006;16:578–585. doi: 10.1016/j.gde.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 46.de Clare M, Pir P, Oliver SG. Haploinsufficiency and the sex chromosomes from yeasts to humans. BMC Biol. 2011;9:15. doi: 10.1186/1741-7007-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ellison CE, et al. Massive changes in genome architecture accompany the transition to self-fertility in the filamentous fungus Neurospora tetrasperma. Genetics. 2011;189(1):55–69. doi: 10.1534/genetics.111.130690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emerson JJ, Kaessmann H, Betrán E, Long M. Extensive gene traffic on the mammalian X chromosome. Science. 2004;303:537–540. doi: 10.1126/science.1090042. [DOI] [PubMed] [Google Scholar]
- 49.Potrzebowski L, et al. Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biol. 2008;6(4):e80. doi: 10.1371/journal.pbio.0060080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herman PK, Rine J. Yeast spore germination: A requirement for Ras protein activity during re-entry into the cell cycle. EMBO J. 1997;16:6171–6181. doi: 10.1093/emboj/16.20.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strathern JN, Herskowitz I. Asymmetry and directionality in production of new cell types during clonal growth: The switching pattern of homothallic yeast. Cell. 1979;17:371–381. doi: 10.1016/0092-8674(79)90163-6. [DOI] [PubMed] [Google Scholar]
- 52.Cliften P, et al. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301(5629):71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- 53.Cliften PF, Fulton RS, Wilson RK, Johnston M. After the duplication: Gene loss and adaptation in Saccharomyces genomes. Genetics. 2006;172:863–872. doi: 10.1534/genetics.105.048900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koren S, Miller JR, Walenz BP, Sutton G. An algorithm for automated closure during assembly. BMC Bioinformatics. 2010;11:457. doi: 10.1186/1471-2105-11-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Byrne KP, Wolfe KH. The Yeast Gene Order Browser: Combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Astell CR, et al. The sequence of the DNAs coding for the mating-type loci of Saccharomyces cerevisiae. Cell. 1981;27(1 Pt 2):15–23. doi: 10.1016/0092-8674(81)90356-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.