Abstract
Most plant species rely on seeds for their dispersal and survival under unfavorable environmental conditions. Seeds are characterized by their low moisture content and significantly reduced metabolic activities. During the maturation phase, seeds accumulate storage reserves and become desiccation-tolerant and dormant. Growth is resumed after release of dormancy and the occurrence of favorable environmental conditions. Here we show that embryonic cotyledon nuclei of Arabidopsis thaliana seeds have a significantly reduced nuclear size, which is established at the beginning of seed maturation. In addition, the chromatin of embryonic cotyledon nuclei from mature seeds is highly condensed. Nuclei regain their size and chromatin condensation level during germination. The reduction in nuclear size is controlled by the seed maturation regulator ABSCISIC ACID-INSENSITIVE 3, and the increase during germination requires two predicted nuclear matrix proteins, LITTLE NUCLEI 1 and LITTLE NUCLEI 2. Our results suggest that the specific properties of nuclei in ripe seeds are an adaptation to desiccation, independent of dormancy. We conclude that the changes in nuclear size and chromatin condensation in seeds are independent, developmentally controlled processes.
Keywords: seed development, seed imbibition, seed germination, chromatin organization
Plant seeds represent a quiescent dehydrated state with significantly reduced metabolic activities, which renders them resistant to abiotic stresses and viable for years. Germination occurs when nondormant seeds meet permissive environmental conditions for humidity, light, and temperature. Seed dormancy evolved to postpone germination during unfavorable seasons (1–3). The moisture content of ripe seeds from the model plant Arabidopsis thaliana is <10% (4). Low humidity levels can cause cellular damage due to protein unfolding and membrane disturbance. Sugars, late embryogenesis abundant proteins, and heat shock proteins play important roles in preventing this damage in seeds (5).
Dehydration, accumulation of storage reserves, and induction of dormancy occur during the seed maturation phase, which is initiated after the embryo has fully developed (4). In Arabidopsis, seed maturation is tightly controlled by at least four central regulators, ABSCISIC ACID-INSENSITIVE 3 (ABI3), LEAFY COTYLEDON 1 (LEC1), LEC2, and FUSCA 3 (FUS3) (3, 6, 7). Other proteins have more dedicated roles in one of these processes; for instance, DELAY OF GERMINATION 1 (DOG1) and HISTONE MONOUBIQUITINATION 1 (HUB1) are required only for the establishment of seed dormancy (8, 9).
Dry seeds represent a transitional state between embryo and seedling. During a phase transition, genes that control the “new” state need to be activated, whereas genes required for the “old” state must be repressed. Chromatin compaction in the cell nucleus has been proposed to contribute to gene regulation by allowing differential accessibility of DNA for the transcription machinery (10, 11). Accordingly, developmental changes in the plant are often accompanied by active modification of the chromatin structure (12). For example, chromatin compaction is loosened in protoplasts and before flowering (13–15), but increases during cell differentiation in maturing leaves and during seedling establishment (10, 16).
Whether the significantly reduced metabolic activity and moisture content of dry seeds are associated with changes in chromatin organization remains unclear, however. Initial observations by Mansfield and Briarty (17) of decreased nuclear volume during seed maturation in Arabidopsis have not been followed up by detailed analyses. Suggestions that chromatin organization is altered during seed development and is genetically controlled come from the identity of HUB1, REDUCED DORMANCY 2 (RDO2), and other factors associated with the polymerase II-associated factor 1 complex (PAF1C). PAF1C modulates the (local) structure of chromatin during transcription elongation, and mutations in factors associated with this complex exhibit reduced seed dormancy (9, 18). In addition, germinating seeds lack conspicuous heterochromatic DNA domains (chromocenters), which reappear during seedling establishment (16).
Here we report a significant decrease in the size of nuclei of embryonic Arabidopsis cotyledons during seed maturation, accompanied by increased chromatin condensation. These properties are reverted during germination. We show that nuclear size reduction and increased chromatin compaction in seeds are independent processes, and propose that they are part of the seed developmental program associated with desiccation tolerance.
Results
Cell Nuclei Decrease in Size During Seed Maturation.
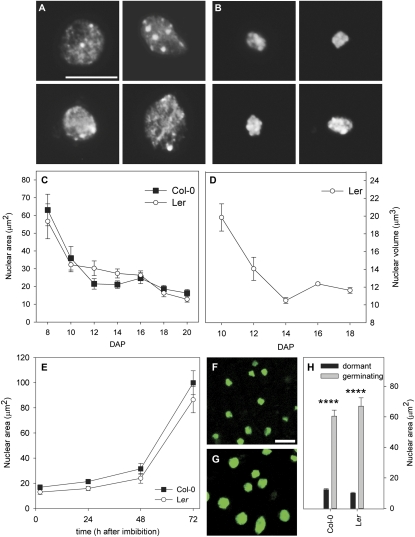
Seed development in Arabidopsis took approximately 20 d under our growth conditions. Embryo development was completed during the first 8–10 d, and the seeds matured during the remainder of the period. We examined the morphology of spread nuclei from embryonic cotyledons during seed maturation in the accessions Columbia-0 (Col) and Landsberg erecta (Ler) using DAPI staining. Strikingly, a significant decrease in nuclear size was observed in embryonic cotyledons during seed maturation (Fig. 1 A and B and Fig. S1). The greatest decrease in size occurred 8–12 d after pollination (DAP) (Fig. 1C). This size decrease was confirmed by nuclear volume measurements (Fig. 1D). The average size of maturing seed nuclei at 10 DAP was 35.9 ± 6.6 μm2 for Col and 32.2 ± 3.9 μm2 for Ler. Interphase nuclei of diploid leaf mesophyll cells of 20-d-old vegetative plants grown simultaneously in the same growth-cabinet were 223.4 ± 29.5 μm2 for Col and 204.1 ± 22.9 μm2 for Ler. This indicates that nuclei in the embryo are already ∼84% smaller than leaf nuclei before the start of seed maturation. Interestingly, the main reduction in nuclear size during maturation was established before major dehydration, which typically occurs between 17 and 20 DAP (4). Adult plant tissues include a range of ploidy levels, and DNA content is positively correlated with nuclear and cell volumes (19). We did not detect major differences in the amount of DAPI staining between nuclei derived from cotyledons at 10 DAP and 20 DAP, indicating that these nuclei contain similar amounts of DNA (Fig. S2). Therefore, differences in ploidy level cannot explain the reduction in nuclear size.
Fig. 1.
Nuclear size dynamics in embryonic cotyledons during seed maturation, imbibition, and germination. (A and B) Representative DAPI-stained embryonic cotyledon nuclei of Col at 10 DAP (A) and 20 DAP (B). (C) Nuclear area during seed maturation of Col (black squares) and Ler (white circles). (D) Kinetics of nuclear volume reduction during seed maturation of Ler. (E) Nuclear area during imbibition and germination of Col (black squares) and Ler (white circles). (F and G) Z-minimum projection-derived confocal microscopy images of 2 h (F) and 72 h (G) imbibed transgenic seeds expressing the nuclear marker Histone 2B fused to fluorescent GFP (Fig. S3). (H) Nuclear areas of cotyledons of 72-h imbibed Col and Ler seeds that were nongerminating (black bars) or germinating (gray bars), derived from the same seed batch. Error bars represent SE; n ≥86 (D; n ≥25), Significance levels: *0.01 > P < 0.05; ****P < 0.0001, two-tailed Student t-test compared with control. (Scale bars: 10 μm.)
Nuclei increased again in size during imbibition/germination (Fig. 1E), as confirmed in vivo using a transgenic line driving the nuclear marker histone H2B fused to fluorescent GFP (Fig. 1 F and G and Fig. S3). Although seeds are saturated with water at 1–2 h after imbibition (20), a significant increase in nuclear size was observed after 24 h of imbibition. This indicates that the seed's hydration status cannot directly explain the dynamics in nuclear size. We studied the relationship between germination and nuclear size by separating germinated seedlings from dormant seeds after a 72-h imbibition of freshly harvested Col and Ler seeds. Cotyledon nuclei from seedlings were significantly (P < 0.0001) larger than those of embryonic cotyledons of dormant seeds (Fig. 1H). This indicates that germination, rather than imbibition, is required for the increase in nuclear size, and that controlling nuclear size is part of the seed development program.
Chromatin in Mature Seeds Is Highly Condensed.
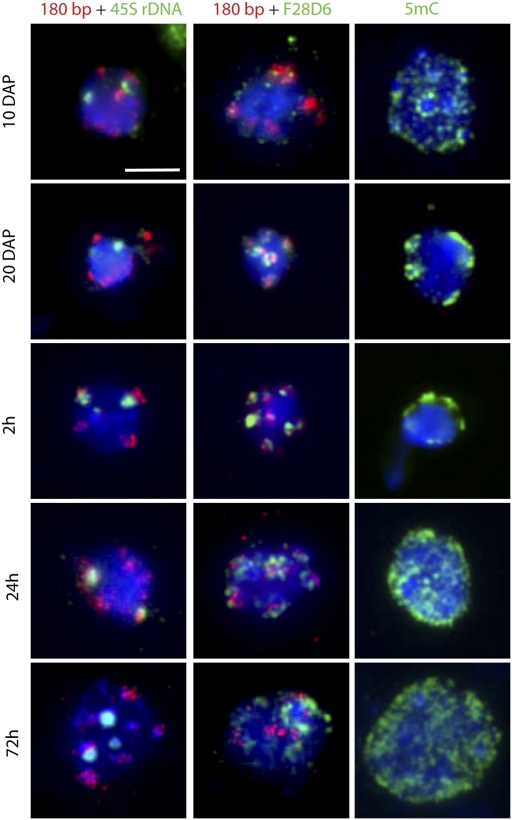
Arabidopsis leaf mesophyll interphase nuclei typically have 6–10 discrete, intensely DAPI-stained “heterochromatic” domains known as chromocenters (10, 21). These domains contain repetitive DNA, such as ribosomal genes, transposable elements, and (peri)centromeric repeats. The small nuclei of embryonic cotyledons in seeds displayed domains with enhanced DAPI staining (Fig. 1 A and B and Fig. S1). More specific staining using Hoechst 33258 dye confirmed these domains as true chromocenters (Fig. S4). We performed FISH experiments to analyze the distribution of centromeric, pericentromeric, and rDNA sequences during seed maturation and imbibition. The 45S rDNA sequences were always condensed, whereas the 180-bp centromeric sequences were condensed during seed maturation but moderately dispersed after 24 h and 72 h after imbibition. The pericentromeric sequences became highly condensed at 20 DAP and 2 h after imbibition, and dispersed after 24 h and 72 h imbibition (Fig. 2 and Fig. S5). Despite the dispersion of centromeric and pericentromeric sequences at 24 h after imibibition, the nucleus was still small at this time (Fig. 1E), indicating that the dynamics of chromatin compaction in seeds is independent of changes in nuclear size.
Fig. 2.
Cytogenetic characterization of embryonic cotyledon nuclei during seed maturation, imbibition, and germination. Representative FISH signals for the centromeric 180-bp repeat (red, Left and Middle) and subtelomeric 45S rDNA repeats (green, Left) or pericentromeric F28D6 (green, Middle) and immunolabeling of 5mC (green, Right) during seed maturation (10 and 20 DAP) and during imbibition/germination (2 h, 24 h, and 72 h). Each nucleus is counterstained with DAPI (blue). (Scale bar: 5 μm.)
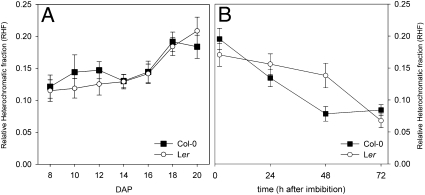
We quantified chromatin condensation by calculating the relative heterochromatic fraction (RHF), an indicator of chromatin compaction in chromocenters (10, 22, 23). A high RHF corresponds to high chromatin compaction. The RHF values in embryonic cotyledons confirmed that chromatin becomes significantly more compact during seed maturation (P = 0.03 in Col and P = 0.003 in Ler, 8 DAP compared with 20 DAP) (Fig. 3A). A maximum RHF value of ∼0.2 was reached in dry seeds. For comparison, young vegetative leaves typically have an RHF of ∼0.08–0.14, whereas fully differentiated leaves have an RHF of ∼0.11–0.16 (10, 24). During imbibition, the RHF again decreased significantly (P = 0.003 in Ler, after 72 h; Fig. 3B) to levels found in leaves and developing embryos. In germinating Col-0 seeds, a significant RHF reduction (P = 0.01) was already observed after 24 h of imbibition, before the increase in nuclear size. This confirms that control of chromatin compaction in seeds is independent of nuclear size.
Fig. 3.
Chromatin compaction in embryonic cotyledon nuclei during seed maturation, imbibition, and germination. RHF during seed maturation (A) and imbibition and germination (B) of Col (black squares) and Ler (white circles). Error bars represent SE; n ≥86.
Because pericentromeric regions are characterized by high levels of methylated DNA, we examined 5-methylcytosine (5mC) patterns at late embryo development (8 DAP) and during seed maturation and germination. We found a striking change in 5mC distribution from early to late seed maturation and during germination. The moderately dispersed 5mC pattern at 8 and 10 DAP became significantly condensed at 20 DAP and 2 h after imbibition, after which a complete dispersion was visible at 24 h and 72 h (Fig. 2 and Fig. S5). These dynamic changes in 5mC distribution reflect the processes of chromatin compaction during seed maturation and decondensation of chromatin during germination. The low-compaction phenotypes at 24 h and 72 h after imbibition resemble those of the decrease in DNA methylation 1 (ddm1) mutant (23). The expression profile of DDM1 cannot account for the observed phenotype, however (Fig. S6).
Dynamics of Nuclear Size Is Regulated by ABI3, LINC1, and LINC2.
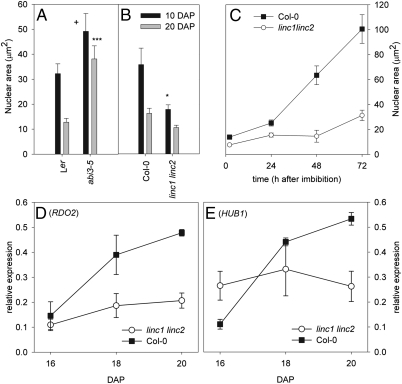
We next studied the influence of the major regulator of seed maturation, ABI3 (7), on nuclear size. One of the most severe mutant alleles, abi3-5, is insensitive to abscisic acid during germination, desiccation-intolerant, and nondormant (25); however, maturing seeds of strong abi3 mutant alleles still have decreased moisture content during seed maturation, only slightly lower than that of WT seeds (26). Nuclear area and volume measurements in abi3-5 embryos during seed maturation revealed larger nuclei and less decrease in size compared with WT at 10 and 20 DAP (Fig. 4A and Fig. S7). WT Ler lost 60.0% of its size during seed maturation (19.5 μm2), compared with a 22.5% (11.1 μm2) loss in abi3-5. This indicates that ABI3 plays a role in controlling nuclear size during seed maturation.
Fig. 4.
ABI3, LINC1, and LINC2 control nuclear size in seeds. (A and B) Nuclear area of embryonic cotyledons of the abi3-5 mutant (Ler) (A) and the double mutant linc1-1 linc2-1 (Col) (B), at 10 DAP (black bars) and 20 DAP (gray bars). Significance levels: +0.05 > P < 0.1; *0.01 > P < 0.05; ***0.0001 > P < 0.001, two-tailed Student t test, compared with control. (C) Nuclear area during imbibition and germination of Col (black squares) and linc1-1 linc2-1 (white circles). Error bars represent SE; n ≥89. (D and E) Relative expression of RDO2 (D) and HUB1 (E) during seed maturation at 16–20 d after pollination compared with ACTIN8 expression. Error bars represent SE; n = 4.
The nuclear matrix constituent protein 1-related nuclear proteins LITTLE NUCLEI 1 (LINC1) and LINC2 are known to affect nuclear morphology in an additive manner, and the double mutant linc1-1 linc2-1 demonstrates a significant reduction in nuclear size and a semidwarf phenotype (27). Nuclei from linc1-1 linc2-1 did not show a significant size decrease during seed maturation. They were already small at 10 DAP and were similar in size to Col WT nuclei at 20 DAP (Fig. 4B). During germination, nuclei size in linc1-1 linc2-1 did not increase significantly, and it remained significantly smaller (0.01 > P < 0.05) compared with Col WT after 72 h (Fig. 4C). This demonstrates that in the absence of LINC1 and LINC2, nuclei remain small (normally seen only in mature seeds) throughout their entire life cycle. FISH and immunolabeling signals from the small linc1-1 linc2-1 nuclei revealed a similar distribution of 180-bp tandem repeats, 45S rDNA, pericentromeric sequences, and 5Mc during imbibition and germination (Fig. S8) as observed for WT (Fig. 2). This indicates that control of chromatin compaction in (germinating) seeds is independent of nuclear size.
We recently reported that the transcription elongation factors HUB1 and RDO2 are up-regulated during seed maturation (18). The hub1-2 and rdo2-1 mutants are characterized by reduced seed dormancy and a high number of differentially expressed genes at the end of seed maturation, including down-regulation of the dormancy gene DOG1 (9, 18). Up-regulation of HUB1 and RDO2 during seed maturation coincides with the decrease in nuclear size, possibly indicating an increased requirement for these factors to maintain transcriptional activity in the small nuclei with enhanced chromatin compaction. In contrast to Col-0, transcript levels of HUB1 and RDO2 did not change between 16 DAP and 20 DAP during seed maturation in the linc1-1 linc2-1 double mutant (Fig. 4 D and E). Moreover, relative HUB1 expression was already greater than that in WT at 16 DAP, suggesting that transcriptional regulation of these transcription elongation factors is controlled by nuclear size and/or LINC1 LINC2.
Nuclear Size Reduction Is Independent of Seed Dormancy and May Be Involved in Desiccation Tolerance.
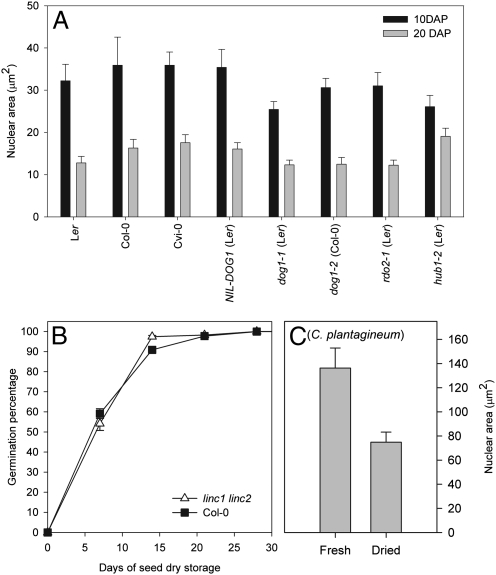
Freshly harvested Arabidopsis seeds are dormant and incapable of germination. The small and highly heterochromatic nuclei of seeds might prevent transcriptional activity and could be part of the dormancy mechanism. Thus, we studied the relationship between dormancy and nuclear size reduction during seed maturation using Arabidopsis genotypes with different dormancy levels. The highly dormant genotypes Cape Verde Islands-0 (Cvi-0) and Near-Isogenic Line NILD106, as well as the nondormant mutants dog1-1 (8), dog1-2, and rdo2-1 (28), showed a similar nuclear size reduction during seed maturation as Ler and Col (Fig. 5A), indicating that seed dormancy is independent of the reduction in nuclear size during seed maturation. In addition, the linc1-1 linc2-1 double mutant with constitutive small nuclei showed similar dormancy levels as Col WT (27) (Fig. 5B). The only exception to this was the nondormant mutant hub1-2 (9) (Fig. 5A), which did not show a significant reduction in nuclear size during seed maturation, similar to abi3-5, indicating that this gene possibly plays a role in controlling nuclear size.
Fig. 5.
Nuclear size reduction is independent of seed dormancy. (A) Nuclear area of embryonic cotyledons of Ler, Col, Cvi-0, NIL-DOG1, dog1-1, dog1-2, rdo2-1, and hub1-2 during seed maturation at 10 DAP (black bars) and 20 DAP (gray bars). (B) Dormancy/germination behavior of Col (black squares) and linc1-1 linc2-1 (white triangles). Germination is expressed as percentage of germinated seeds after different periods of seed dry storage (days). (C) Nuclear area of fresh and dehydrated C. plantagineum leaves. Error bars represent SE; n ≥102 (B, n ≥23).
The low moisture content of ripe Arabidopsis seeds (<10%; ref. 4) suggests that the reduction in nuclear size and increased chromatin condensation might be an adaptation to very low moisture contents. We tested this hypothesis by measuring the nuclear size in leaves of the desiccation-tolerant resurrection plant Craterostigma plantagineum (29). We found that desiccation of leaves led to a significant decrease in nuclear size (Fig. 5C), suggesting that the decreased nuclear size is not restricted to seeds, but could represent a universal mechanism in acquisition of desiccation tolerance.
Discussion
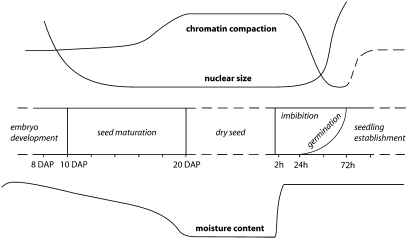
Nuclei in the embryonic cotyledon of dry seeds are significantly reduced in size and have highly condensed chromatin. These properties are established during seed maturation and reversed during germination, as shown schematically in Fig. 6. The decrease in nuclear size is independent of the reduction in cell size and ploidy levels, because the cotyledon cell volume even increases during embryo development and early seed maturation (4, 17). We found no major differences in ploidy levels in nuclei at 10 DAP and 20 DAP (Fig. S2).
Fig. 6.
Schematic representations of the dynamics of chromatin compaction, nuclear size, and moisture content during the transition from embryo to seedling. Arabidopsis embryonic cotyledon nuclei significantly decrease in size during seed maturation and increase again during germination (Fig. 1). Chromatin compaction increases throughout seed maturation, resulting in a high level of compaction in dry seeds (Figs. 2 and 3). In the first 3 d after imbibition, the chromatin decondensates dramatically, followed by a rapid increase in compaction during early seedling establishment and vegetative growth. Note that the dynamics of chromatin compaction and nuclear size are independent processes that occur largely independent of changes in moisture content. The dynamics of moisture content during seed maturation is based on Baud et al. (4), and that during imbibition is based on Preston et al. (20). Chromatin compaction during seedling establishment is based on Mathieu et al. (16). The y-axes represent relative scales.
Why Are Nuclei in Ripe Seeds Compact?
Observations in germinating and dormant Phaseolus vulgaris seeds have demonstrated that nuclei reversibly shrink toward the dormant phase (30). In addition, studies of Phaseolus lunatus and maize (Zea mays) found smaller nuclei in dormant seeds compared with germinated seeds, suggesting that chromatin is more compact in dormant seeds (31). Using genotypes with differing dormancy levels, we have shown here that the nuclear size of embryonic cotyledons is independent of seed dormancy in Arabidopsis. Mansfield and Briarty (17) suggested that a reduction in nuclear size could be related to lower transcription rates and water loss in seeds. Analogously, reduced nuclear size has been reported in petioles of the desert plant Zygophyllum dumosum during the dry season (32), and we found decreased nuclear size in C. plantagineum leaves after desiccation. In Arabidopsis, the main decrease in nuclear size during seed maturation occurs between 10 and 14 DAP (Fig. 6), with only minor decreases thereafter. This develops in concert with the establishment of desiccation tolerance, which occurs between 13 and 15 DAP (33) and precedes the major decrease in water content, from ∼32% to <10%, occurring between 17 and 20 DAP (4). Nuclear size also is not directly influenced by the initial uptake of water in dry seeds during imbibition (Fig. 1 E and H and Fig. 6). ABI3 is required for the nuclear size reduction during seed maturation, and LINC1 and LINC2 are required for the size increase during germination. Taken together, these findings suggest that the decrease in nuclear size is an active process that is likely part of a general mechanism for establishing desiccation tolerance in plants.
Transition from Embryo to Seedling.
Developmental phase transitions are generally characterized by a transient loosening of chromatin compaction (10–16). In contrast, we found increased chromatin compaction in the seed during the transition from embryo to seedling (Fig. 6). This indicates that the transitional state of seeds deviates from other phase transitions, probably because seeds represent a prolonged quiescent state under desiccated conditions. Our FISH and 5mC immunolabeling results demonstrate that chromatin decondensation does occur during imbibition of dry seeds and is independent of the changes in nuclear size (Fig. 2 and Fig. S8). This suggests that the transition from embryo to seedling at the chromatin level occurs during imbibition, as also proposed by Comai and Harada (34) based on the timing of transcriptional activities of germination specific genes.
Influence of Chromatin Compaction on Nuclear Functioning.
To the best of our knowledge, the reduced nuclear size in Arabidopsis is specific for (maturing) seeds and might not be adequate for full nuclear functioning in other tissues. In accordance with this, the linc1-1 linc2-1 double mutant with constitutive small nuclei shows a semidwarf and leaf-curling phenotype (27). A nucleus of a given size cannot contain an unlimited amount of DNA; Fujimoto et al. (35) reported an upper limit of 3% for the ratio of DNA volume to nuclear volume by comparing root tip cells across plant species with different genome sizes. In Arabidopsis embryonic cotyledon nuclei with reduced size at 20 DAP, this ratio does not exceed 1%, which is well below the upper limit found in plants but above the value of 0.26% reported by Fujimoto et al. (35) in Arabidopsis root tip cells. This confirms that the chromatin in these reduced nuclei is highly compacted.
Increased chromatin compaction is associated with decreased transcriptional activity (10–12). Consistently, active transcription in mature seeds is very low or absent, although run-on studies have shown that several genes remain transcriptionally competent in dry seeds (34). We have shown that nuclei regain their normal size and structure only after germination (Fig. 6), suggesting that transcription rates are fully restored only after germination. This implies that the germination process itself does not require high transcription rates. In accordance with this finding, it has been shown that germination can occur without active transcription but requires translation, probably from mRNAs stored in the seed (36).
The situation is different during seed maturation, when nuclei are small and the chromatin is highly condensed, although active transcription of maturation-specific genes is still required. We recently reported that genes encoding PAF1C factors are up-regulated toward the end of seed maturation, and that mutants of these genes have reduced dormancy (18). PAF1C provides a platform for the association of complexes that modulate the (local) structure of chromatin during transcription elongation (37). Our data indicate that at least two of these PAF1C-associated factors, HUB1 and RDO2, were not up-regulated during seed maturation in the linc1 linc2 double mutant with constitutively small nuclei. HUB1 expression was already increased before the end of seed maturation in these small nuclei. Interestingly, HUB1 and RDO2 homologs were also identified as being up-regulated during desiccation in a transcriptome analysis of C. plantagineum (38). We propose that up-regulation of PAF1C-associated factors represents a general mechanism for maintaining gene expression in dehydrating nuclei with decreased size and increased chromatin compaction.
Materials and Methods
Plant Materials and Growth Conditions.
The rdo2-1 (Ler) mutant (28), hub1-2 (Ler) (9), NILD106 (containing the Cvi-0 DOG1 allele in the Ler isogenic background) (39), dog1-1 (8), linc1-1 linc2-1 (Col) (27), and abi3-5 (Ler) (25, 40) have been described previously. All of these mutants were kind gifts of the authors who described the lines. The dog1-2 (Col) mutant, containing a premature stop codon, was isolated by V. Raz (Wageningen University, Wageningen, The Netherlands).
Plants were grown in soil containing a mixture of substrate and vermiculite (3:1) in growth cabinets (Elbanton) in 16 h light at 22 °C and 8 h dark at 16 °C. Seeds for imbibition and germinating experiments, nuclear size/volume measurements, FISH, and immunolabeling were sown on filter paper in Petri dishes, placed into transparent moisturized containers, and incubated in a germination cabinet (Van den Berg Klimaattechniek) in 16 h light at 25 °C and 8 h dark at 20 °C.
C. plantagineum was grown in an artificial clay substrate as described previously (38). Leaves of 6- to 8-wk-old plants were either fixed directly in Carnoy fixative or obtained from plants that had been dehydrated by withholding water to reach a relative water content <5%.
Germination Tests.
Approximately 50 seeds of individually harvested plants were sown on filter paper and incubated in a germination cabinet as described above. After 7 d of incubation, the germination percentages were determined. After-ripening of dry seeds was performed in darkness at 21 °C and 50% relative humidity in a controlled-atmosphere cabinet (MMM Medcenter).
Cytogenetic Experiments.
Tissues were fixed in Carnoys fixative, except those for volume measurements, which were fixed in 1% formaldehyde. Embryonic cotyledons from intact seeds and seedlings were isolated manually using dissection needles under a binocular and were separated from hypocotyl and root tissue before further processing and analysis.
Spread preparations of nuclei were made as described previously (22, 24, 41) and in SI Materials and Methods. Nuclear size (area of the spread nucleus) and RHF [fluorescence intensity of intensely DAPI-stained chromocenters, relative to the fluorescence of the entire nucleus (22)] measurements were performed using a macro (42) in ImagePro-Plus (Media Cybernetics). Each data point comprises ≥86 nuclei of ≥100 pooled embryonic cotyledons derived from two biological replicates, each consisting of at least five plants (Table S1). Nuclear volumes were measured from Z-stacked confocal images as described previosuly (43), using Imaris software (Bitplane). GFP signals from the transgenic line pH2B:H2B::GFP were detected by Z-minimum projection using a Zeiss TCS SP2 confocal microscope.
FISH and immunolabeling experiments were carried out as described previously (22, 24, 41). Further details are provided in SI Materials and Methods.
Quantitative RT-PCR.
Seed RNA was extracted following an adapted protocol of the RNAqueous small-scale phenol-free total RNA isolation kit with an RNA isolation aid (Ambion), as described previously (18). cDNA synthesis was performed with the QuantiTect Reverse Transcription Kit (Qiagen) including DNase treatment. Quantitative RT-PCR was performed using the QuantiTect SYBR Green PCR Kit (Qiagen). Expression was calculated relative to ACT8 (AT1G49240), which has been shown to be an appropriate reference gene for seed-derived cDNA (44). Primers are listed in Table S2.
Supplementary Material
Acknowledgments
We thank Penka Pavlova (Wageningen University) for kindly sharing the nuclear area measurement macro and Eric J. Richards (Boyce Thompson Institute for Plant Research) for providing the linc1-1 linc2-1 mutant and the Centre for Advanced Microscopy (CAM) Amsterdam for using the microscope facility. This work was supported by an EMBO Long-Term Fellowship (ALTF 700-2010, to M.v.Z.) and grants from the Netherlands Organization for Scientific Research (ALW2PJ/09053, to M.A.K. and P.F.), the Deutsche Forschungsgemeinschaft (SO 691/3-1, to W.J.J.S.), and the Max Planck Society (to W.J.J.S. and M.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117726108/-/DCSupplemental.
References
- 1.Finch-Savage WE, Leubner-Metzger G. Seed dormancy and the control of germination. New Phytol. 2006;171:501–523. doi: 10.1111/j.1469-8137.2006.01787.x. [DOI] [PubMed] [Google Scholar]
- 2.Finkelstein R, Reeves W, Ariizumi T, Steber C. Molecular aspects of seed dormancy. Annu Rev Plant Biol. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- 3.Holdsworth MJ, Bentsink L, Soppe WJJ. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008;179:33–54. doi: 10.1111/j.1469-8137.2008.02437.x. [DOI] [PubMed] [Google Scholar]
- 4.Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C. An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem. 2002;40:151–160. [Google Scholar]
- 5.Hoekstra FA, Golovina EA, Buitink J. Mechanisms of plant desiccation tolerance. Trends Plant Sci. 2001;6:431–438. doi: 10.1016/s1360-1385(01)02052-0. [DOI] [PubMed] [Google Scholar]
- 6.Santos-Mendoza M, et al. Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 2008;54:608–620. doi: 10.1111/j.1365-313X.2008.03461.x. [DOI] [PubMed] [Google Scholar]
- 7.To A, et al. A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell. 2006;18:1642–1651. doi: 10.1105/tpc.105.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bentsink L, Jowett J, Hanhart CJ, Koornneef M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:17042–17047. doi: 10.1073/pnas.0607877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Koornneef M, Soppe WJJ. The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell. 2007;19:433–444. doi: 10.1105/tpc.106.049221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tessadori F, van Driel R, Fransz P. Cytogenetics as a tool to study gene regulation. Trends Plant Sci. 2004;9:147–153. doi: 10.1016/j.tplants.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 11.Fransz P, de Jong H. From nucleosome to chromosome: A dynamic organization of genetic information. Plant J. 2011;66:4–17. doi: 10.1111/j.1365-313X.2011.04526.x. [DOI] [PubMed] [Google Scholar]
- 12.Exner V, Hennig L. Chromatin rearrangements in development. Curr Opin Plant Biol. 2008;11:64–69. doi: 10.1016/j.pbi.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Tessadori F, et al. Large-scale dissociation and sequential reassembly of pericentric heterochromatin in dedifferentiated Arabidopsis cells. J Cell Sci. 2007a;120:1200–1208. doi: 10.1242/jcs.000026. [DOI] [PubMed] [Google Scholar]
- 14.Tessadori F, Schulkes RK, van Driel R, Fransz P. Light-regulated large-scale reorganization of chromatin during the floral transition in Arabidopsis. Plant J. 2007b;50:848–857. doi: 10.1111/j.1365-313X.2007.03093.x. [DOI] [PubMed] [Google Scholar]
- 15.Grafi G, et al. Histone methylation controls telomerase-independent telomere lengthening in cells undergoing dedifferentiation. Dev Biol. 2007;306:838–846. doi: 10.1016/j.ydbio.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 16.Mathieu O, et al. Changes in 5S rDNA chromatin organization and transcription during heterochromatin establishment in Arabidopsis. Plant Cell. 2003;15:2929–2939. doi: 10.1105/tpc.017467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mansfield SG, Briarty LG. Cotyledon cell development in Arabidopsis thaliana during reserve deposition. Can J Bot. 1992;70:151–164. [Google Scholar]
- 18.Liu Y, et al. Identification of the Arabidopsis REDUCED DORMANCY 2 gene uncovers a role for the polymerase-associated factor 1 complex in seed dormancy. PLoS ONE. 2011;6:e22241. doi: 10.1371/journal.pone.0022241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jovtchev G, Schubert V, Meister A, Barow M, Schubert I. Nuclear DNA content and nuclear and cell volume are positively correlated in angiosperms. Cytogenet Genome Res. 2006;114:77–82. doi: 10.1159/000091932. [DOI] [PubMed] [Google Scholar]
- 20.Preston J, et al. Temporal expression patterns of hormone metabolism genes during imbibition of Arabidopsis thaliana seeds: A comparative study on dormant and non-dormant accessions. Plant Cell Physiol. 2009;50:1786–1800. doi: 10.1093/pcp/pcp121. [DOI] [PubMed] [Google Scholar]
- 21.Fransz P, De Jong JH, Lysak M, Castiglione MR, Schubert I. Interphase chromosomes in Arabidopsis are organized as well-defined chromocenters from which euchromatin loops emanate. Proc Natl Acad Sci USA. 2002;99:14584–14589. doi: 10.1073/pnas.212325299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tessadori F, et al. Phytochrome B and histone deacetylase 6 control light-induced chromatin compaction in Arabidopsis thaliana. PLoS Genet. 2009;5:e1000638. doi: 10.1371/journal.pgen.1000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soppe WJJ, et al. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 2002;21:6549–6559. doi: 10.1093/emboj/cdf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Zanten M, et al. Photoreceptors CRYTOCHROME2 and phytochrome B control chromatin compaction in Arabidopsis. Plant Physiol. 2010;154:1686–1696. doi: 10.1104/pp.110.164616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ooms J, Leon-Kloosterziel KM, Bartels D, Koornneef M, Karssen CM. Acquisition of desiccation tolerance and longevity in seeds of Arabidopsis thaliana (a comparative study using abscisic acid-insensitive abi3 mutants) Plant Physiol. 1993;102:1185–1191. doi: 10.1104/pp.102.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nambara E, Keith K, McCourt P, Naito S. A regulatory role for the ABI3 gene in the establishment of embryo maturation in Arabidopsis thaliana. Development. 1995;121:629–636. [Google Scholar]
- 27.Dittmer TA, Stacey NJ, Sugimoto-Shirasu K, Richards EJ. LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. Plant Cell. 2007;19:2793–2803. doi: 10.1105/tpc.107.053231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peeters AJM, et al. Characterization of mutants with reduced seed dormancy at two novel rdo loci and a further characterization of rdo1 and rdo2 in Arabidopsis. Physiol Plant. 2002;115:604–612. doi: 10.1034/j.1399-3054.2002.1150415.x. [DOI] [PubMed] [Google Scholar]
- 29.Bartels D, Salamini F. Desiccation tolerance in the resurrection plant Craterostigma plantagineum: A contribution to the study of drought tolerance at the molecular level. Plant Physiol. 2001;127:1346–1353. [PMC free article] [PubMed] [Google Scholar]
- 30.Kater JM. A cytological study of dormancy in the seed of Phaseolus vulgaris. Ann Bot (Lond) 1927;41:629–641. [Google Scholar]
- 31.Middendorf FG. Cytology of dormancy in Phaseolus and Zea. Bot Gaz. 1939;100:485–499. [Google Scholar]
- 32.Granot G, et al. Histone modifications associated with drought tolerance in the desert plant Zygophyllum dumosum Boiss. Planta. 2009;231:27–34. doi: 10.1007/s00425-009-1026-z. [DOI] [PubMed] [Google Scholar]
- 33.Koornneef M, Hanhart CJ, Hilhorst HWM, Karssen CM. In vivo inhibition of seed development and reserve protein accumulation in recombinants of abscisic acid biosynthesis and responsiveness mutants in Arabidopsis thaliana. Plant Physiol. 1989;90:463–469. doi: 10.1104/pp.90.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Comai L, Harada JJ. Transcriptional activities in dry seed nuclei indicate the timing of the transition from embryogeny to germination. Proc Natl Acad Sci USA. 1990;87:2671–2674. doi: 10.1073/pnas.87.7.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujimoto S, Ito M, Matsunaga S, Fukui K. An upper limit of the ratio of DNA volume to nuclear volume exists in plants. Genes Genet Syst. 2005;80:345–350. doi: 10.1266/ggs.80.345. [DOI] [PubMed] [Google Scholar]
- 36.Rajjou L, et al. The effect of α-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol. 2004;134:1598–1613. doi: 10.1104/pp.103.036293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez MC, et al. Transcriptomes of the desiccation-tolerant resurrection plant Craterostigma plantagineum. Plant J. 2010;63:212–228. doi: 10.1111/j.1365-313X.2010.04243.x. [DOI] [PubMed] [Google Scholar]
- 39.Alonso-Blanco C, Bentsink L, Hanhart CJ, Blankestijn-de Vries H, Koornneef M. Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics. 2003;164:711–729. doi: 10.1093/genetics/164.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bies-Etheve N, et al. Importance of the B2 domain of the Arabidopsis ABI3 protein for Em and 2S albumin gene regulation. Plant Mol Biol. 1999;40:1045–1054. doi: 10.1023/a:1006252512202. [DOI] [PubMed] [Google Scholar]
- 41.Schubert I, Fransz PF, Fuchs J, de Jong JH. Chromosome painting in plants. Methods Cell Sci. 2001;23:57–69. [PubMed] [Google Scholar]
- 42.Pavlova P, Tessadori F, de Jong HJ, Fransz P. Immunocytological analysis of chromatin in isolated nuclei. Methods Mol Biol. 2010;655:413–432. doi: 10.1007/978-1-60761-765-5_28. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, Geyer R, Brambilla V, Nakabayashi K, Soppe WJJ. Chromatin dynamics during seed dormancy. Methods Mol Biol. 2011;773:239–257. doi: 10.1007/978-1-61779-231-1_15. [DOI] [PubMed] [Google Scholar]
- 44.Graeber K, Linkies A, Wood AT, Leubner-Metzger G. A guideline to family-wide comparative state-of-the-art quantitative RT-PCR analysis exemplified with a Brassicaceae cross-species seed germination case study. Plant Cell. 2011;23:2045–2063. doi: 10.1105/tpc.111.084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.