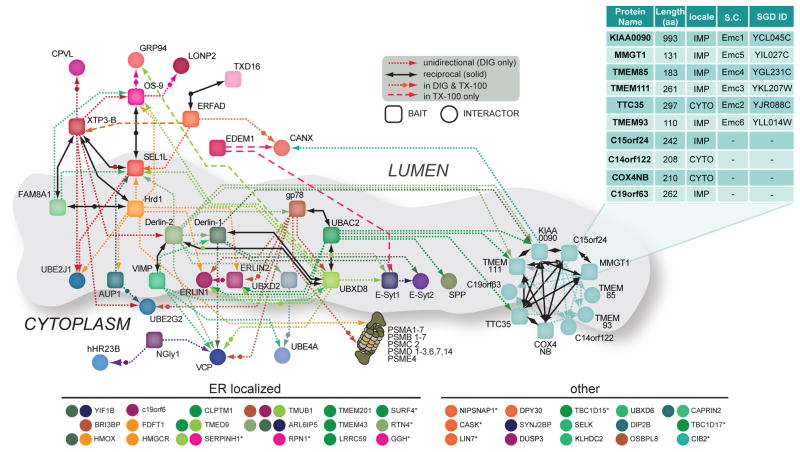
Figure 2.
The Interaction Network for ERAD (INfERAD). Interaction network for ERAD isolated in DIG and TX-100 represented by baits (squares) and their HCIPs (circles). Unidirectional (short dash, single arrow) and reciprocal (solid black, double arrows) interactions are shown. Each bait protein is rendered in a unique colour and line colour reflects the bait protein used to identify the interaction with the HCIP. Short dashed lines marked with a circle indicate interactions detected in both DIG and TX-100, while long dashed lines represent those found only in TX-100. The inset table lists the determined constituents of the mEMC, their size, cellular localisation, and corresponding yeast orthologs (SC) and ID in the Saccharomyces Genome Database (SGD). For clarity, a selection of additional DIG HCIPs not included in the map is shown on the bottom, with a circle’s colour corresponding to the bait for which the HCIP was observed and asterisks denoting an HCIP also detected in TX-100.