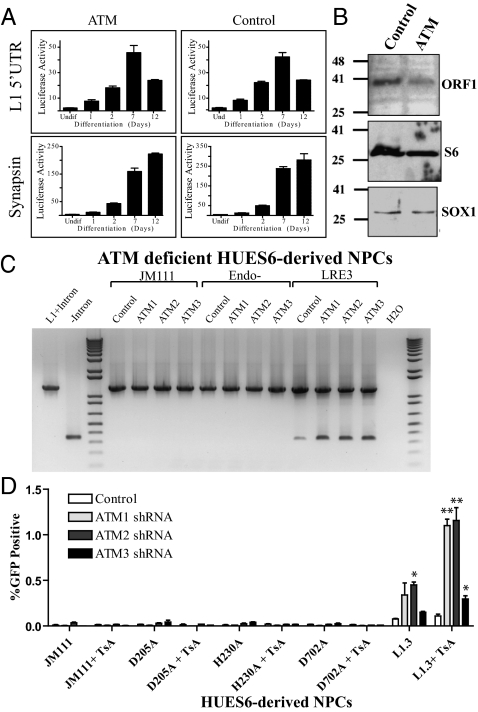
Fig. 3.
Examination of L1 retrotransposition in ATM-deficient HUES6-derived NPCs. (A) The L1 5′ UTR is rapidly induced upon HUES6-derived NPC differentiation in both ATM-deficient and ATM-proficient shRNA-infected cells, as is synapsin, a marker of differentiation. The x axis indicates days postdifferentiation; the y axis indicates luciferase fold activity. (B) RNP particles were isolated from control, and ATM-deficient NPCs and were subjected to Western blotting using anti-ORF1p and antiribosomal S6 antibodies. SOX1 is a whole-cell lysate control. (C) PCR of genomic DNA confirmed L1 retrotransposition. The 1,243-bp product corresponds to the EGFP retrotransposition indicator cassette (lane 1, L1 + intron); the 342-bp product is diagnostic for intron loss (lane 2, control EGFP no intron). (D) L1 retrotransposition (EGFP-positive cells) is detected in HUES6-derived NPCs transfected with a WT L1 (L1.3–EGFP), but not in cells transfected with mutant L1 constructs [L1.3/JM111–EGFP (ORF1 mutant), L1.3/D205A–EGFP, or L1.3/H230A–EGFP (endonuclease mutants) or L1.3/D702A–EGFP (reverse transcriptase mutant)]. Notably, L1.3–EGFP retrotransposition is modestly increased in ATM-deficient cells compared with controls. Also, the addition of the histone deacetylase inhibitor trichostatin A (TsA) enhanced the ability to detect retrotransposition. Error bars indicate SEM. *P < 0.05 and **P < 0.01 are a result of a one-way ANOVA with Bonferroni correction.