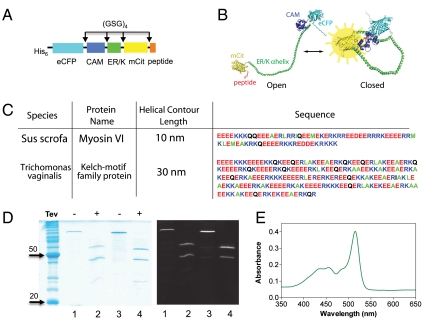
Fig. 2.
SPASM sensor design. (A) Schematic of protein domains (His6 is at the N terminus) in the SPASM sensor. A 12 amino acid Gly-Ser-Gly (GSG)4 linker is placed between different domains to ensure rotational freedom. (B) Structural model of the SPASM sensor in the open (Left) and closed (Right) conformation. (C) Sequences of two ER/K α-helices. Note that the 20-nm helix used in this study is the first 130 amino acids of the 30-nm helix sequence. (D, Left) Coomassie gel staining of purified SPASM sensors with 30 nm (lanes 1 and 2) and 20 nm (lanes 3 and 4) ER/K α-helices. Lanes 2 and 4 were treated with TEV-protease, which cleaves at its recognition site engineered between the CAM and ER/K α-helix. (Right) Laser fluorescence gel scan (457-nm eCFP excitation with 520BP40 emission). This condition captures both eCFP and mCitrine fluorescent bands. (E) Absorption spectrum of purified SPASM sensor showing peaks at 433 nm (eCFP) and 514 nm (mCit). Note that mCit has approximately twofold higher extinction coefficient compared to eCFP.