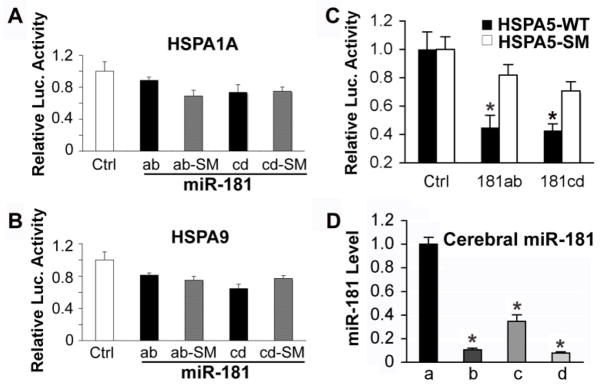
Fig. 1.
Hspa5 is the target of miR-181. A, B. Dual luciferase activity assays performed in BOSC23 cells co-transfected with the plasmid containing Renilla luciferase followed by the Hspa1a or Hspa9 3′UTR (WT) and plasmids encoding either pri-miR-181ab or pri-miR-181cd or their seed mutants (SM) demonstrates that miR-181 does not recognize either of these 3′UTRs, as there is no reduction of luciferase activity compared to the SM control. C. The same assay performed with the Hspa5 3′UTR (Hspa5-WT) or its seed mutant (Hspa5-SM) shows that miR-181ab and cd both reduce luciferase activity. *P<0.05, statistically different from the SM group by T-test. Luciferase assays were performed 3 times in triplicate. D. Differing levels of expression of miR-181a, b, c, and d are detected in cortex of normal control brains. N=4 animals, assayed in triplicate, * P<0.01 different than levels of miR-181a by ANOVA and Newman-Keuls post hoc test.