Abstract
The species-rich family Mycosphaerellaceae contains considerable morphological diversity and includes numerous anamorphic genera, many of which are economically important plant pathogens. Recent revisions and phylogenetic research have resulted in taxonomic instability. Ameliorating this problem requires phylogenetic placement of type species of key genera. We present an examination of the type species of the anamorphic Asperisporium and Pantospora. Cultures isolated from recent port interceptions were studied and described, and morphological studies were made of historical and new herbarium specimens. DNA sequence data from the ITS region and nLSU were generated from these type species, analysed phylogenetically, placed into an evolutionary context within Mycosphaerellaceae, and compared to existing phylogenies. Epitype specimens associated with living cultures and DNA sequence data are designated herein. Asperisporium caricae, the type of Asperisporium and cause of a leaf and fruit spot disease of papaya, is closely related to several species of Passalora including P. brachycarpa. The status of Asperisporium as a potential generic synonym of Passalora remains unclear. The monotypic genus Pantospora, typified by the synnematous Pantospora guazumae, is not included in Pseudocercospora sensu stricto or sensu lato. Rather, it represents a distinct lineage in the Mycosphaerellaceae in an unresolved position near Mycosphaerella microsora.
Keywords: Ascomycota, Capnodiales, Dothideomycetes, lectotype, pawpaw, Pseudocercospora ulmifoliae
INTRODUCTION
Mycosphaerella and related fungi are classified in Capnodiales (Dothideomycetes, Ascomycota) and include thousands of species (Crous et al. 2007). These fungi have diverse ecological roles, especially as saprophytes and parasites, and numerous species are of agricultural significance (Crous 2009). As plant pathogens, these fungi are found on plant taxa across the embryophytes (Farr & Rossman 2011) with most species exhibiting host specificity (Crous 2009).
In terms of morphology, generic concepts for Mycosphaerella and its related anamorphs have been challenging as the number of variable and overlapping characters has led to different classifications that emphasize different characters (Baker et al. 2000, Crous & Braun 2003, Crous et al. 2007, 2009b). With progress towards phylogenetic hypotheses based on DNA sequence data, it has become apparent that older morphological classifications are riddled with non-monophyletic groups and unexpected bedfellows (Crous et al. 2007, 2009b, Crous 2009).
The backbone of our understanding of the phylogeny of this group is skewed in large part, but not entirely, towards sampling from fungi associated with hosts in two plant families, Myrtaceae and Proteaceae (Crous et al. 2007, 2009a, b, Crous 2009). Furthermore, the number of genetic loci that have been examined is relatively small. While the type species of Mycosphaerella, M. punctiformis, has been placed in a phylogenetic context (Verkley et al. 2004), type species of large, related genera have yet to be placed (Crous & Braun 2003, Crous et al. 2007, 2009b).
During the course of conducting work related to plant protection and quarantine, we obtained new collections via port interceptions that were identified as Asperisporium caricae and Pantospora guazumae. These collections represent the generic types of Asperisporium (Ellis 1971) and Pantospora (Deighton 1976), respectively. Asperisporium is a small genus that includes roughly 12 species (Kirk et al. 2008). It shares many morphological features with Passalora such as pigmented conidia and thickened and darkened conidiogenous loci while it is differentiated by verrucose conidia (Crous & Braun 2003, Schubert & Braun 2005), but this distinction has been considered doubtful (Crous & Braun 2003, Schubert & Braun 2005). Asperisporium caricae is responsible for an important leaf and fruit spot disease of Carica papaya (papaw or papaya) (Stevens 1939) that is commonly referred to as black spot, blight or ‘rust’ of pawpaw (Ellis & Holliday 1972). The synnematous Pantospora is a monotypic genus that causes a leaf spot disease of Guazuma ulmifolia (Deighton 1976). Deighton (1976) and Crous & Braun (2003) both noted its similarity to Pseudocercospora, especially in regards to the dictyospores in the type species of Pseudocercospora, P. vitis, and the latter authors formerly classified it in that genus. Since neither Asperisporium nor Pantospora have been placed phylogenetically (Crous & Braun 2003, Schubert & Braun 2005), we generated DNA sequence data from the ITS and nLSU, conducted analyses, present phylogenetic placements of these genera, and discuss them in the context of existing phylogenetic studies (Crous et al. 2007, 2009b). To further stabilize the application of Asperisporium and Pantospora, we herein epitypify the type species of these genera with collections associated with living cultures and DNA sequence data. Morphological descriptions of the designated epitypes and associated ex-epitype cultures are presented, and historical collections and descriptions were studied to confirm conspecificity of the new collections.
MATERIALS AND METHODS
Morphology and herbarium material
Dried herbarium material was rehydrated and viewed in 3 % KOH (Largent et al. 1977), and microscopic observations of cultures were made of material mounted in 3 % KOH or buffered Shear’s mounting fluid (Graham 1959). Herbarium acronyms follow Thiers (2011). See Farr & Rossman (2011) for additional information about collections housed at BPI.
Cultures
Isolates in pure culture were grown in plastic Petri plates on 2 % Difco potato-dextrose agar (PDA) and BBL Sabouraud dextrose agar (SDA), which were both prepared according to the manufacturers’ instructions. Cultures were incubated at 24 °C with a 12 h light/dark regimen. A subset of these cultures was transferred to a 12 h black light/dark regimen also at 24 °C after approximately 2–2.5 wk to promote sporulation. Sporulation was also promoted by exposing 1 mo old cultures that were otherwise incubated in the dark at ambient room temperature to 1 h of UV at an intensity setting of 60 with a Fisher Biotech Transilluminator FBTIV-816 at approximately 2–3 d intervals. Terminology for colour includes general terms from author notes as well as standard terminology with the sample reference code in parentheses from Kornerup & Wanscher (1967). Voucher cultures were deposited at the Centraalbureau voor Schimmelcultures (CBS).
DNA extraction, PCR amplification, and sequencing
DNA was extracted from fresh mycelium using the Qiagen DNeasy Plant Mini Kit (Gaithersburg, Maryland). DNA sequence data were generated from the nuclear encoded ribosomal ITS region (ITS1, 5.8S, ITS2), the nuclear encoded ribosomal large subunit (28S nLSU), and a portion between amino acid motifs 5–7 of the nuclear DNA-dependant RNA Polymerase II gene’s large subunit (RPB2) for Asperisporium caricae and Pantospora guazumae. PCR cocktails for all reactions contained 0.2 μM of each forward and reverse primer, GoTaq Flexi Buffer (Promega; Madison, Wisconsin), 0.2 mM dNTPs, 2.0 mM Mg2+, and 5U/μL Pomega GoTaq. Primers used for PCR and sequencing were fRPB2-5F and fRPB2-7cR (Liu et al. 1999) for RPB2, ITS5 and ITS4 (White et al. 1990) for the ITS, and LROR (Monclavo et al. 2000) and LR7 (Vilgalys & Hester 1990) for the nLSU. Thermal cycling conditions for RPB2 and nLSU were those of Malkus et al. (2006) and Reeb et al. (2004), respectively. Thermal cycling conditions for the ITS were: 95 °C for 60 s; 35 cycles at 95 °C for 15 s, 55 °C for 20 s, and 72 °C for 1 min; and a final extension at 72 °C for 3 min. Cycle sequencing was conducted using BigDye v. 3.1 (Applied Biosystems; Foster City, California) sequencing chemistry. Resulting fluorescent-labelled fragments were sequenced on an ABI 3730 capillary sequencer. Electropherograms were edited in the program Geneious Pro v. 5 (Drummond et al. 2010). Sequences were submitted to GenBank (http://www.ncbi.nlm.nih.gov).
Data matrix and phylogenetic analysis
For the purpose of determining preliminary phylogenetic positions of A. caricae and P. guazumae among members of Mycosphaerellaceae and related genera, their nLSU sequences were manually incorporated into the alignment of Crous et al. (2009b) and analyzed phylogenetically using Maximum Parsimony (MP) as the optimality criterion in the program TNT v. 1.1 (Goloboff et al. 2008).
Based on these results (not presented), a selection of taxa was made to provide the appropriate phylogenetic context for a fine-scale placement of these taxa within Mycosphaerellaceae. Additional taxa were included based on similarity and determined using BLAST results of Genbank. In all cases, the ITS and the nLSU sequences retrieved from GenBank to represent a particular taxon and combined for analysis were generated from the same culture.
Multiple sequence alignment of the ITS and the nLSU was conducted within the program Geneious Pro v. 5 (Drummond et al. 2010) using MUSCLE v. 3.6 (Edgar 2004), and adjusted manually. Insertions and deletions (indels) within the concatenated ITS and nLSU matrix were coded using the ‘simple indel coding’ method of Simmons & Ochoterena (2000) as implemented in Gapcode.py (Ree 2007). The resulting alignment was deposited into TreeBASE as accession number SN11747. Bayesian Inference (BI) phylogenetic analysis was conducted on the concatenated ITS/nLSU matrix, including indel characters, in MrBayes v. 3.1.2 (Huelsenbeck & Ronquist 2001, Ronquist & Huelsenbeck 2003). Indel data were analyzed as ‘restriction’ data type and DNA sequence data were analyzed as ‘DNA’ data type. The GTR+I+G model of DNA sequence evolution was determined as the best fit using the Akaike Information Criterion (AIC; Posada & Buckley 2004) in MrModeltest v. 2.2 (Nylander 2004) and implemented in the BI analysis. The coding parameter for the restriction data type was set to variable. All other parameters were left as default. The posterior probability (pp) distribution of trees was estimated based on the results of two independent runs of 1 million generations each of the Markov Chain Monte Carlo (MCMC) simulation, which sampled trees every 100 generations until the standard deviation of split frequencies reached less than 0.01. The burn-in was determined using the program Tracer v. 1.5 (Rambaut & Drummond 2007). The remaining trees were combined and used to build a 50 % majority rule consensus within the program FigTree v. 1.3.1 (Rambaut 2009).
RESULTS
Data matrix and phylogenetic analysis
The combined ITS, nLSU, plus indel matrix contained 1 304 characters, 53 of which were indels. Of these 1 251 nucleotide characters, 202 were variable (16.1 %) and 126 were informative (10.1 %). Thirty-one indel characters were informative (58.5 %). The ITS alignment was 504 nucleotide positions in length, 130 of which were variable (25.8 %) and 81 informative (16.1 %). The nLSU alignment was 747 nucleotide positions, 72 of which were variable (9.6 %) and 45 informative (6.0 %). The ITS alignment contributed 46 indels, 28 of which were informative (60.9 %). The nLSU alignment contributed seven indels, three of which were informative (42.9 %).
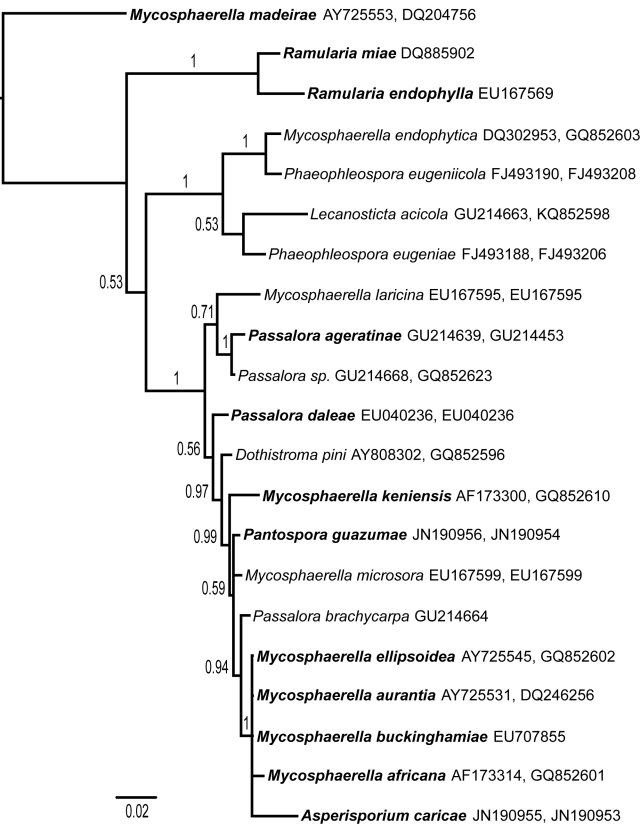
Preliminary MP analysis of the nLSU suggested that A. caricae and P. guazumae occupy phylogenetic positions within the clade with 95 % bootstrap support (Crous et al. 2009b) containing Dothiostroma (clade 7), which is sister to the clade comprised of Pseudocercospora-like fungi (clade 6), Phaeophleospora (clade 5), and Lecanosticta (clade 4). Our combined analysis of ITS, nLSU, and indels resulted in strong support (pp = 1.0) for this clade and an increased level of resolution among its members relative to nLSU alone (Fig. 1). Within this clade, P. guazumae occupies an unresolved position with Mycosphaerella microsora sister to a well-supported clade (p = 0.94) containing Passalora brachycarpa, M. ellipsoidea, M. aurantia, M. buckinghamiae, M. africana, and A. caricae. The latter four species are members of a well-supported (pp = 1.0) polytomy sister to Passalora brachycarpa.
Fig. 1.
A 50 % majority rule Bayesian Inference phylogram resulting from analysis of the ITS, the nLSU, and associated indels. The scale bar is proportional to the amount of character evolution on the tree. The ITS and nLSU Genbank accession numbers are provided on terminals, in that order, after each taxon name. Only a single accession is labelled on terminals where the Genbank record contains concatenated ITS and nLSU sequences. Taxon names in bold indicate ex-types.
TAXONOMY
Asperisporium Maubl., Lavoura 16: 207/212. 1913 ‘1912’, and Bull. Trimestriel Soc. Mycol. France 29: 357. 1913. Note: Maublanc published his article in two languages in Lavoura and separately in Bull. Trimestriel Soc. Mycol. France. We were unable to confirm which printed issuance was first.
Typus genericus. Asperisporium caricae (Speg.) Maubl.
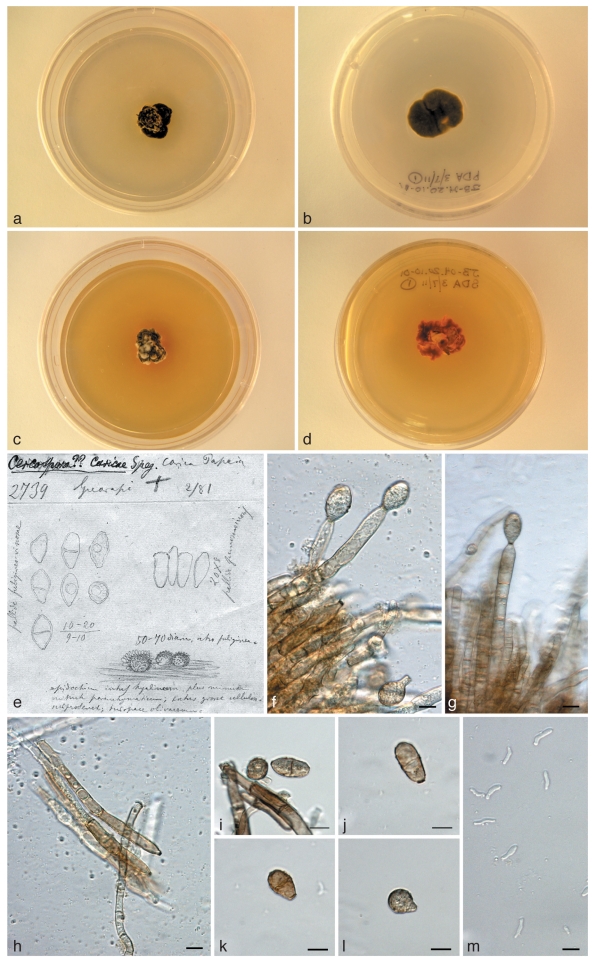
Asperisporium caricae (Speg.) Maubl., Lavoura 16: 207/212. 1913 ‘1912’, and Bull. Trimestriel Soc. Mycol. France 29: 357. 1913. — Fig. 2
Fig. 2.
Cultures and microscopic features of Asperisporium caricae. a–d. Ex-epitype (CBS 130298) at approximately 1 mo at 24 °C with a 12 h light/dark regimen: a. PDA; b. reverse on PDA; c. SDA; d. reverse on SDA. — e. Lectotype packet, No. 2739 (LPS). — f–m. Ex-epitype (CBS 130298) on SDA; f–h. conidiophores and conidia; i–l. conidia; m. spermatia. — Scale bars = 10 μm for all.
Basionym. Cercospora caricae Speg., Anales Soc. Ci. Argent. 22: 215. 1886. Note: This species was published as ‘Cercospora? caricae’ to indicate doubt as to the generic classification, but it is still valid according to ICBN Art. 34.1 (McNeill et al. 2006).
≡ Fusicladium caricae (Speg.) Sacc., Rend. Congr. Bot. Palermo: 58. 1902.
[≡ Pucciniopsis caricae (Speg.) Höhn., Centralbl. Bakteriol., 2. Abth.: 60: 5. 1923, nom. illeg., non Earle 1902. Isonym (Speg.) Seaver, Scientific Survey of Porto Rico and the Virgin Islands, Vol. VIII, Part 1: 104. 1926.] Note: This combination was published as ‘Pucciniopsis? caricae’ to indicate doubt as to the generic classification, but it is still valid according to ICBN Art. 34.1.
= Scolicotrichum caricae Ellis & Everh., J. Mycol. 7: 134. 1892 as ‘Scolecotrichum’.
= Epiclinium cumminsii Massee, Bull. Misc. Inform. Kew 1898: 133. 1898.
= Pucciniopsis caricae Earle, Bull. New York Bot. Gard. 2: 340. 1902.
Types and typifications
Lectotypus of Cercospora caricae designated by Chupp, A Monograph of the Fungus Genus Cercospora: 106. 1953: Paraguay, Guarapi, on leaves of Carica papaya, Feb. 1881, coll. B. Balansa, No. 2739 (LPS).
Epitypus of Cercospora caricae hic designatus: Brazil, Intercepted at USA, Washington, Seattle, entering from Brazil, on fruit of Carica papaya, 16 Apr. 2010, coll. C. Weight, isolated by J.F. Bischoff from BPI 880773, epitype is a dried culture on SDA (BPI 881135); ex-epitype CBS 130298; GenBank accession nos: ITS (JN190955), LSU (JN190953), RPB2 (JN190951).
Notes — Spegazzini (1886) cited two collections, no. 2739 and no. 3855, in the protologue without indicating either as type. These are syntypes according to ICBN Art. 9.4. Chupp (1953) lectotypified the species via ICBN Art. 7.11 when he indicated the word type for no. 2739 at LPS.
Description of the epitype (preserved culture) and collection from which it was isolated
Fructicolous with spots scattered, 3–4 mm diam. Sporodochia formed on stromata, immersed becoming erumpent, punctiform, blackish to black. In culture on SDA, sporodochia of loosely to densely arranged conidiophores produced on darkly pigmented stromata. Conidiophores macronematous, mononematous, simple or less commonly branched, more or less straight to slightly sinuous, smooth, brownish, septate, 58–168 × 5–10 μm. Conidiogenous cells integrated, terminal, polyblastic, sympodial, cylindrical to clavate, at times slightly geniculate, conidiogenous loci thickened, darkened. Conidia 14–22 × 8–13 μm, solitary, broadly ellipsoid, ellipsoid, obovate, pyriform, or oblong and somewhat clavate, smooth becoming verrucose, hyaline becoming brownish, each typically 0–1-septate with one more or less median septum, rarely with 2 septa, hila thickened, darkened, occasionally on an apiculus-like base. Spermogonia with spermatogenous cells cylindrical, lageniform, or ampulliform, smooth, hyaline. Spermatia 6–10 × 1–2.5 μm, solitary, cylindrical, clavate, ellipsoid, sigmoid, or stocking-shaped, apices obtuse, bases truncate, smooth, hyaline, aseptate, at times with a frill-like structure below basal septum.
Culture characteristics — Colonies on PDA 9–10 mm after 1 mo at 24 °C with a 12 h light/dark regimen; mycelium forming a raised mound composed of tiers of smaller mounds, surface slightly velutinous, near dark green (30F3–4), at times portions covered with whitish, short erect hyphae or whitish aerial hyphae; with scattered black spherical structures, spermogonia, 80–160 μm diam; margin lobed, whitish to concolorous; reverse concolorous. Surface mycelium with hyphae branching, walls smooth, hyaline to brownish, septate, 3–6.5 μm diam. Sporulation within approximately 1 mo, but precise time not noted. Conidial production on PDA sparse in comparison to that on SDA.
Colonies on SDA 8–9 mm after 1 mo at 24 °C with a 12 h light/dark regimen; mycelium forming a raised mound composed of tiers of smaller mounds, surface velutinous, near dark green (30F3) to greenish grey (30F2), at times portions covered with whitish, short erect hyphae, dense lanose to cottony whitish hyphae, or whitish aerial hyphae; with scattered black spherical structures, spermogonia, 80–160 μm diam; margin lobed, more or less concolorous; reverse near brownish orange (6C7). Surface mycelium with hyphae branching, walls smooth, hyaline to brownish, septate, 3–6.5 μm diam. Sporulation confirmed at 3 wk.
Colonies on both media showing slightly increased growth rates on a 12 h black light/dark regimen. Sporulation more or less as above. Colonies exposed to UV light via the transilluminator showed increased growth rate as submerged, uncoloured hyphae with a corresponding lack of mycelium above the agar surface.
Habitat & Distribution — Leaves and fruits of Carica papaya, rarely Carica chilensis (≡ Vasconcellea chilensis) (Caricaceae) (Ellis & Holliday 1972, Crous & Braun 2003). This fungus is widely distributed in subtropical and tropical regions with reports from Africa, Asia, North America, Oceania, and South America (Crous & Braun 2003, Farr & Rossman 2011).
Specimens examined. Bermuda, Paget, Agricultural Station, on leaves of Carica papaya, 24 June 1921, coll. W.H. Whetzel (BPI 424787). – Brazil, Intercepted at USA, Washington, Seattle, entering from Brazil, on fruit of Carica papaya, 16 Apr. 2010, coll. C. Weight (BPI 880773); culture isolated by J.F. Bischoff from BPI 880773, dried culture on SDA (BPI881135, designated epitype). – Colombia, Intercepted at New York, J.F.K.I.A., #24116, on fruit of Carica papaya, 4 Jan. 1969, coll. C. Smock (BPI 424779). – Cuba, Santiago de Las Vegas, Experiment Station, on leaves of Carica papaya, 8 May 1915, coll. R.A. Jehle (BPI 424780). – Dominican Republic, Valle del Cibao, Santiago Prov., Santiago, Hato del Yaque, Gardens, on leaves of Carica papaya, 19 Jan. 1931, coll. R. Ciferri, Mycoflora Domingensis Exsiccata 311 (BPI 424783). – Honduras, Jamastran, on leaves of Carica papaya, 10 Mar. 1964, coll. A.S. Muller (BPI 424789). – Mexico, Intercepted at Texas, Laredo, #008382, on fruit of Carica papaya, 20 Feb. 1975, coll. S. Kendall (BPI 424778). – Puerto Rico, Yauco, on leaves of Carica papaya, 30 Mar. 1916, coll. W.H. Whetzel and E.W. Olive (BPI 424777). – Venezuela, Federal District, Valle de Puerto La Cruz, El Limón, on leaves of Carica papaya, 15 Jan. 1925, coll. H. Sydow (BPI 1112163).
Notes — The collection from which the epitype was isolated is a cut portion of fruit. Unfortunately, it could not be well preserved before it was overgrown with anamorphic fungi including Penicillium. Thus, we have designated a dried culture isolated from this collection as the epitype. Conidiophores in culture were longer than those observed on host tissues (Ellis 1971, Ellis & Holliday 1972).
Ellis & Holliday (1972) listed three taxonomic synonyms. Examination of the protologues of these names supports the synonymy. Maublanc (1913a, b) described a teleomorph that he suggested was associated with Asperisporium caricae as Sphaerella caricae. This morph was later classified as Mycosphaerella caricae (Maubl.) Hansf., which makes it an illegitimate later homonym of Mycosphaerella caricae Syd. & P. Syd. These teleomorphic names have been considered to be conspecific and associated with the anamorphic Phoma caricae (Sivanesan 1984). Others expressed doubt that A. caricae has a known teleomorph since the connection has never been proven (Crous & Braun 2003). Recently, M. caricae Syd. & P. Syd. and M. caricae (Maubl.) Hansf. were listed separately with each having different anamorphs, but the type of the latter was not studied (Aptroot 2006). Mycosphaerella caricae Syd. & P. Syd. was transferred to Stagonosporopsis with no mention of the later homonym in the synonym list (Aveskamp et al. 2010). No additional information about the life cycle of A. caricae in regards to its ascal state and the synonymy of these historical names can be added by this study. However, a structure of unproven identity that is presumed to be a spermogonium was found in culture. Its presumptive spermatogenous cells and spermatia are reminiscent of those described by Crous (1998) for Mycosphaerella crystallina. The presumptive spermatia did not germinate on PDA. The presence of spermogonia suggests that A. caricae has a sexual stage in its life cycle.
Pantospora Cif., Ann. Mycol. 36: 242. 1938.
Typus genericus. Pantospora guazumae Cif.
= Dictyocephala A.G. Medeiros, Universidade do Recife, Instituto de Micologia, Publicação No. 372: 13. 1962.
Typus genericus. Cercospora ulmifoliae Obreg.-Bot., which was indirectly referenced via the invalid Dictyocephala ulmifoliae (Obreg.-Bot.) A.G. Medeiros (ICBN Art. 10.3).
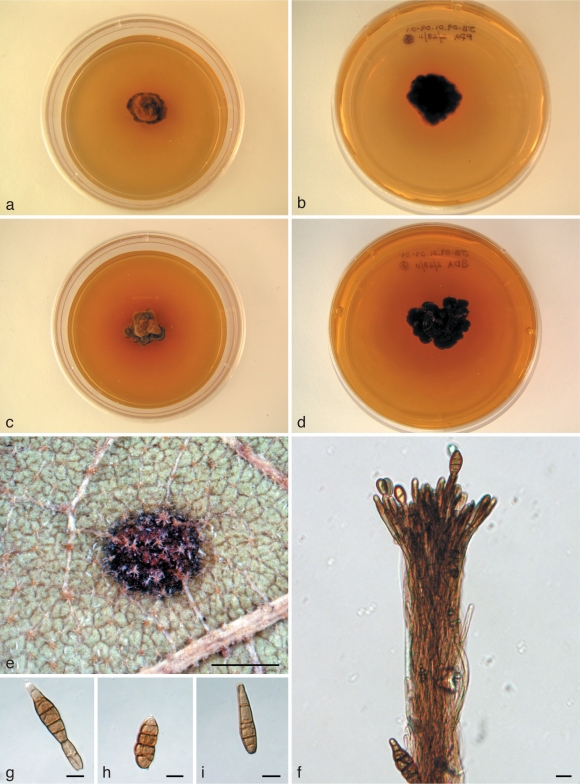
Pantospora guazumae Cif., Ann. Mycol. 36: 242. 1938. — Fig. 3
Fig. 3.
Cultures and microscopic features of Pantospora guazumae. a–d. Ex-epitype (CBS 130299) at approximately 1 mo at 24 °C with a 12 h light/dark regimen: a. PDA; b. reverse on PDA; c. SDA; d. reverse on SDA. — e. Leaf spot on abaxial surface of Guazuma ulmifoliae (BPI 880778, designated epitype); f. synnema on Guazumae ulmifoliae (BPI 880778, designated epitype); g–i. conidia from ex-epitype (CBS 130299) on PDA. — Scale bars = 1 mm for e, 10 μm for f–i.
= Cercospora ulmifoliae Obreg.-Bot., Caldasia 1: 51. 1941.
≡ Dictyocephala ulmifoliae (Obreg.-Bot.) A.G. Medeiros, Universidade do Recife, Instituto de Micologia, Publicação No. 372: 13. 1962 as ‘ulmifolii’, nom. inval. via ICBN Art. 33.4.
≡ Pseudocercospora ulmifoliae (Obreg.-Bot.) U. Braun & Crous, CBS Biodiversity Series 1: 415. 2003. Note: Pseudocercospora guazumae (Syd.) Deighton prevented a legitimate combination based on Pantospora guazumae (Crous & Braun 2003).
Types and typifications
Lectotypus of Pantospora guazumae designated by Deighton, Mycol. Pap. 140: 159. 1976: Dominican Republic, Valle del Cibao, prov. Santiago, Hato del Yaque, on leaves of Guazuma ulmifolia, 20 Apr. 1930, coll. R. Ciferri & A.M. Borgna Ciferri, Batey no. 1, Mycoflora Domingensis Exsiccata 210 (IMI 59269, K(M) 169346).
Epitypus of Pantospora guazumae hic designatus: Mexico, Intercepted at USA, Arizona, Nogales, entering from Mexico, on leaf of Guazuma ulmifolia, 12 Feb. 2009, coll. J. Moore (BPI 880778); ex-epitype CBS 130299; GenBank accession nos: ITS (JN190956), LSU (JN190954), RPB2 (JN190952).
Notes — Ciferri (1938) distributed the original material as number 210 in exsiccatae sets. These are syntypes according to ICBN Art. 9.4. Deighton (1976) lectotypified the species when he indicated that the collection at IMI was the type. This specimen is now housed at K.
Description of the epitype
Leaf spots scattered, 1.5–2 mm diam, visible on both adaxial and abaxial surfaces, typically circular, dark brown to blackish with a distinct, lighter, occasionally purplish, margin on adaxial surface, discoloured brown with a distinct, lighter, occasionally purplish, margin on abaxial surface. Caespituli hypophyllous, scattered within margin of leaf spots, conidiophores densely aggregated forming synnemata. Synnemata basistromatic with immersed stromata, erect, more or less even with hyphal tips spreading apart at apex, up to 290 μm long, up to 40 μm wide along non-apical portion. Conidiophores unbranched, more or less straight to somewhat sinuous with apices more or less obtuse, often interwoven, smooth, pale brown to brown, septate, 3–6 μm, widest towards synnematal apex. Conidiogenous cells integrated, terminal, blastic, slightly verrucose and with annellations, conidiogenous loci visible, not significantly thickened or darkened. Conidia up to 43 × 13 μm, solitary, versiform, ellipsoid with short beaks to obclavate, slightly verrucose, brown, with multiple transverse, longitudinal, and occasionally oblique septa, hila visible but not thickened or darkened.
Culture characteristics — Colonies on PDA 12–13 mm after 1 mo at 24 °C with a 12 h light/dark regimen; mycelium forming a raised mound composed of tiers of smaller mounds, surface lanose, near brownish orange (7C7) to light brown (7D7); margin lobed, more or less concolorous; reverse almost black or black; medium becoming discoloured with a greyish red (7B6) to orange (6B7) soluble pigment. Surface mycelium with hyphae branching, walls smooth, at times covered with orange crystals, hyaline to brownish orange in Shear’s mounting fluid, purple in 3 % KOH, septate, 5–6.5 μm diam. Sporulation not observed.
Colonies on SDA 11–13 mm after 1 mo at 24 °C with a 12 h light/dark regimen; mycelium forming a raised mound composed of tiers of smaller mounds, surface lanose, multicoloured with near brownish orange (7C7) to light brown (7D4) to orangish tan to greyish hues; margin lobed, more or less concolorous; reverse almost black or black; medium becoming discoloured with a greyish red (7B6) to orange (6B7) soluble pigment. Surface mycelium with hyphae branching, walls smooth, at times covered with orange, pigmentary crystals, hyaline, orangish to brownish orange in Shear’s mounting fluid, purple in 3 % KOH, septate, 5–6.5 μm diam. Sporulation not observed.
Colonies on both media showing increased growth rates on a 12 h black light/dark regimen. Sporulation not observed. Colonies exposed to UV via the transilluminator showed increased growth rate, a significant increase in submerged hyphae with dark pigmentation at margins with a corresponding lack of mycelium above, and production of a few scattered synnemata. Mature synnemata were observed approximately 1.5 wk after the UV regimen was initiated. Synnemata in culture were of greater width and fascicles of conidiophores were somewhat more loosely arranged than on host leaves. Conidia 32–60 × 8–16 μm, versiform, ellipsoid with short beaks, obclavate, cylindrical-obclavate, or more or less cylindrical, slightly verrucose, brown, with multiple transverse, longitudinal, and occasionally oblique septa. Both dictyosporous and scolecosporous conidia present, additional longitudinal and/or oblique septa may develop over time. Otherwise, synnemata similar in culture and on host leaves.
Habitat & Distribution — Leaves of Guazuma ulmifolia (Malvaceae). According to Farr & Rossman (2011), this fungus is known from North America (Cuba, Dominican Republic) and South America (Brazil, Colombia). This is the first report of this species from Mexico.
Specimens examined. Colombia, Rioclaro, near Cali, on Guazuma ulmifolia, June 1938, coll. C. Garces-Orejuela (BPI 445535). – Dominican Republic, Valle del Cibao, prov. Santiago, Hato del Yaque, on leaves of Guazuma ulmifolia, 20 Apr. 1930, coll. R. Ciferri & A.M. Borgna Ciferri, Batey no. 1, Mycoflora Domingensis Exsiccata 210 (IMI 59269, K(M) 169346, lectotype); (BPI 445536, syntype); Feb. 1932, coll. R. Ciferri (BPI 445537); 01 Sept. 1931, coll. R. Ciferri (BPI 445538); 01 Sept. 1931, coll. R. Ciferri (BPI 445539). – Mexico, Intercepted at USA, Arizona, Nogales, entering from Mexico, on leaf of Guazuma ulmifolia, 12 Feb. 2009, coll. J. Moore (BPI 880778, designated epitype).
Notes — This epitype description is based on sparse, dried herbarium material. The epitype specimen is not fully mature as it possesses small leaf spots and lacks a large number of mature conidia. In the interest of preserving the specimen, the sample size of measured structures is relatively small. Discrepancies with the observations of previous authors should not be considered significant. As with and loosely following Medeiros (1962), UV was found to stimulate the production of synnemata in culture.
Both Deighton (1976) and Crous & Braun (2003) considered Cercospora ulmifoliae as a synonym of Pantospora guazumae even though they did not study Obregón-Botero’s original material of C. ulmifoliae. Based on the descriptions provided by Obregón-Botero (1941) and Chupp (1953), there can be no doubt that the two names are synonymous. There is no known sexual stage for Pantospora guazumae.
DISCUSSION
Asperisporium has been thought of as a likely synonym of Passalora since the two genera were separated on the basis of conidial surface ornamentation (Crous & Braun 2003, Schubert & Braun 2005). These authors tentatively maintained them as distinct due to the absence of DNA sequence data. The phylogenetic analyses place Asperisporium caricae, the type of the genus, in a relatively close relationship with several species of Passalora including P. brachycarpa. Sequence data are available for only a small number of the approximately 580 species of Passalora (Kirk et al. 2008) and the type species of the genus, Passalora bacilligera, has not been sequenced and placed phylogenetically. As Passalora currently stands, it is a polyphyletic genus (Crous et al. 2009b). A final conclusion cannot be made on the status of Asperisporium. The collective works of Patil & Thirumalachar (1966), Ellis (1976), Barreto & Evans (1995), Braun (2000a, b) and Braun & Crous (2007) cover nearly all of the remaining species that are currently classified in Asperisporium.
Crous & Braun (2003) formally classified Pantospora guazumae in Pseudocercospora, noted the lack of molecular data for it and explained why the presence of dictyospores may not be a distinguishing character at the rank of genus. Based on the phylogenetic analyses, Pantospora is not closely related to two clades presented by Crous et al. (2009b), namely, Pseudocercospora including the type species, P. vitis (clade 16) and Pseudocercospora-like (clade 14). Pantospora is in an unresolved position near Mycosphaerella microsora. Pantospora appears to be a distinct lineage.
In summary, the type species of Asperisporium, A. caricae, and Pantospora, P. guazumae, have been placed phylogenetically in the Mycosphaerellaceae. Studies of their culture characteristics were made and both species were epitypified with herbarium material associated with living cultures and DNA sequence data. The phylogenetic placement of these genera demonstrates that previous generic concepts and the perceived values of particular morphological features were not necessarily congruent with phylogeny. Since this is becoming a repeated result (Crous & Braun 2003, Crous et al. 2007, 2009b, Crous 2009) and that sampling in terms of taxa and numbers of genetic loci remains small for such a large family, we advocate a conservative and patient approach towards the creation of new taxonomic schemes and nomenclatural novelties in this group of fungi.
Acknowledgments
We thank Jorge A. Chayle and other staff at LPS for providing information about and images of the original material of Asperisporium caricae and permission to reproduce the image of the lectotype packet. Curators at K are acknowledged for their loan of the lectotype of Pantospora. Pedro W. Crous and Uwe Braun are thanked for their comments about preliminary phylogenetic trees and generic concepts. Amy Y. Rossman provided helpful comments to improve the manuscript prior to its submission.
REFERENCES
- Aptroot A.2006. Mycosphaerella and its anamorphs: 2. Conspectus of Mycosphaerella. CBS Biodiversity Series 5: 1– 231 [Google Scholar]
- Aveskamp M, Gruyter H de, Woudenberg J, Verkley G, Crous PW.2010. Highlights of the Didymellaceae: A polyphasic approach to characterize Phoma and related pleosporalean genera. Studies in Mycology 65: 1– 60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker WA, Partridge EC, Morgan-Jones G.2000. Notes on hyphomycetes. LXXVIII. Asperisporium sequoiae, the causal organism of conifer needle blight, reclassified in Cercosporidium, with comments on the status of the genus. Mycotaxon 76: 247– 256 [Google Scholar]
- Barreto RW, Evans HC.1995. The mycobiota of the weed Mikania micrantha in southern Brazil with particular reference to fungal pathogens for biological control. Mycological Research 99: 343– 352 [Google Scholar]
- Braun U.2000a. Miscellaneous notes on some micromycetes. Schlechtendalia 5: 31– 56 [Google Scholar]
- Braun U.2000b. Annotated list of Cercospora spp. described by C. Spegazzini. Schlechtendalia 5: 57– 79 [Google Scholar]
- Braun U, Crous PW.2007. The diversity of cercosporoid hyphomycetes – new species, combinations, names and nomenclatural clarifications. Fungal Diversity 26: 55– 72 [Google Scholar]
- Chupp C.1953. A monograph of the fungus genus Cercospora. Published by the author, Ithaca, New York [Google Scholar]
- Ciferri R.1938. Mycoflora domingensis exsiccata. Annales Mycologici 36: 198– 245 [Google Scholar]
- Crous PW.1998. Mycosphaerella spp. and their anamorphs associated with leaf spot diseases of Eucalyptus. Mycological Memoirs 21: 1– 170 [Google Scholar]
- Crous PW.2009. Taxonomy and phylogeny of the genus Mycosphaerella and its anamorphs. Fungal Diversity 38: 1– 24 [Google Scholar]
- Crous PW, Braun U.2003. Mycosphaerella and its anamorphs: 1. Names published in Cercospora and Passalora. CBS Biodiversity Series 1: 1– 571 [Google Scholar]
- Crous PW, Braun U, Groenewald JZ.2007. Mycosphaerella is polyphyletic. Studies in Mycology 58: 1– 32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Carnegie AJ, Wingfield MJ, Groenewald JZ.2009a. Novel species of Mycosphaerellaceae and Teratosphaeriaceae. Persoonia 23: 119– 146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Carnegie AJ, Wingfield MJ, Hunter GC, Burgess TI, Andjic V, Barber PA, Groenewald JZ.2009b. Unravelling Mycosphaerella: do you believe in genera? Persoonia 23: 99– 118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deighton FC.1976. Studies on Cercospora and allied genera. VI. Pseudocercospora Speg., Pantospora Cif. and Cercoseptoria Petr. Mycological Papers 140: 1– 168 [Google Scholar]
- Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Heled J, Kearse M, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A.2010. Geneious v. 5.1, Available from www.geneious.com
- Edgar RC.2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792– 1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MB.1971. Dematiaceous hyphomycetes. Commonwealth Mycological Institute, Kew, Surrey, England: [Google Scholar]
- Ellis MB.1976. More dematiaceous hyphomycetes. Commonwealth Mycological Institute, Kew, Surrey, England: [Google Scholar]
- Ellis MB, Holliday P.1972. Asperisporium caricae. CMI Descriptions of Pathogenic Fungi and Bacteria 347: 1– 2 [Google Scholar]
- Farr DF, Rossman AY.2011. Fungal databases, systematic mycology and microbiology laboratory, ARS, USDA. Retrieved March 30, 2011 from nt.ars-grin.gov/fungaldatabases/
- Goloboff PA, Farris JS, Nixon KC.2008. TNT, a free program for phylogenetic analysis. Cladistics 24: 774– 786 [Google Scholar]
- Graham SO.1959. The effects of various reagents, mounting media, and dyes on the teliospore walls of Tilletia controversa Kühn. Mycologia 51: 477– 491 [Google Scholar]
- Huelsenbeck JP, Ronquist F.2001. MrBayes: Bayesian inference of phylogeny. Bioinformatics 17: 754– 755 [DOI] [PubMed] [Google Scholar]
- Kirk PM, Cannon PF, Minter DW, Stalpers JA. with assistance. 2008. Ainsworth & Bisby’s dictionary of the fungi, tenth edition. Cab International, Wallingford, UK: [Google Scholar]
- Kornerup A, Wanscher JH.1967. Methuen handbook of color, 2nd ed Methuen & Co Ltd, London, UK: [Google Scholar]
- Largent D, Johnson D, Watling R.1977. How to identify mushrooms to genus III. Microscopic features. Mad River Press, Eureka, CA: [Google Scholar]
- Liu YJ, Whelen S, Hall BD.1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Molecular Biology and Evolution 16: 1799– 1808 [DOI] [PubMed] [Google Scholar]
- Malkus A, Chang P-FL, Sabina MZ, Chung K, Shao J, Cunfer BM, Arseniuk E, Ueng PP.2006. RNA polymerase II gene (RPB2) encoding the second largest protein subunit in Phaeosphaeria nodorum and P. avenaria. Mycological Research 110: 1152– 1164 [DOI] [PubMed] [Google Scholar]
- Maublanc A.1913a ‘1912’. Sobre uma molestia do mamoeiro (Caryca Papayal, L.)/Sur une maladie des feuilles du papayer “Carica papaya”. Lavoura 16: 204– 212 [Google Scholar]
- Maublanc A.1913b. Sur une maladie des feuilles du papayer (Carica papaya). Bulletin Trimestriel de la Société Mycologique de France 29: 353– 358 [Google Scholar]
- McNeill J, Barrie FR, Burdet HM, Demoulin V, Hawksworth DL, Marhold K, Nicolson DH, Prado J, Silva PC, Skog JE, Wiersema JH, Turland NJ. (eds). 2006. International code of botanical nomenclature (Vienna code): Adopted by the Seventeenth International Botanical Congress, Vienna, Austria, July 2005. Gantner, Ruggell, Liechtenstein: [Google Scholar]
- Medeiros AG.1962. Dictyocephala novo género de fungos Dematiaceae. Universidade do Recife, Instituto de Micologia, Publicação No. 372: 1– 24 [Google Scholar]
- Monclavo JM, Lutzoni FM, Rehner SA, Johnson J, Vilgalys R.2000. Phylogenetic relationships of agaric fungi based on nuclear large subunit ribosomal DNA sequences. Systematic Biology 49: 278– 305 [DOI] [PubMed] [Google Scholar]
- Nylander JAA.2004. MrModeltest, v. 2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University: [Google Scholar]
- Obregón-Botero R.1941. Cuatro nuevos deuteromicetos Colombianos. Caldasia 1: 49– 51 [Google Scholar]
- Patil BV, Thirumalachar MJ.1966. Studies on some fungi of Masharashtra-India-I. Sydowia 20: 33– 38 [Google Scholar]
- Posada D, Buckley TR.2004. Model selection and model averaging in phylogenetics: advantages of akaike information criterion and Bayesian approaches over likelihood ratio tests. Systematic Biology 53: 793– 808 [DOI] [PubMed] [Google Scholar]
- Rambaut A.2009. FigTree v. 1.3.1. Computer program and documentation distributed by the author at http://tree.bio.ed.ac.uk/software/
- Rambaut A, Drummond AJ.2007. Tracer v. 1.5. Computer program and documentation distributed by the authors at http://beast.bio.ed.ac.uk/Tracer [Google Scholar]
- Ree RH.2007. Gapcode.py, v. 2.0. Distributed by the author at http://www.reelab.net/home/ [Google Scholar]
- Reeb V, Lutzoni F, Roux C.2004. Multilocus phylogenetic circumscription of the lichen-forming fungi family Acarosporaceae and its position within the Ascomycota. Molecular Phylogeny and Evolution 32: 1036– 1060 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP.2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572– 1574 [DOI] [PubMed] [Google Scholar]
- Schubert K, Braun U.2005. Taxonomic revision of the genus Cladosporium s.l. 4. Species reallocated to Asperisporium, Dischloridium, Fusicladium, Passalora, Pseudoasperisporium and Stenella. Fungal Diversity 20: 187– 208 [Google Scholar]
- Simmons MP, Ochoterena R.2000. Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology 49: 369– 381 [PubMed] [Google Scholar]
- Sivanesan A.1984. The bitunicate ascomycetes and their anamorphs. Gantner Verlag, Vaduz, Liechtenstein: [Google Scholar]
- Spegazzini C.1886. Fungi Guaranitici. Pugillus I. Anales de Sociedad Científica Argentina 22: 186– 224 [Google Scholar]
- Stevens HE.1939. Papaya diseases. Proceedings of the Florida State Horticultural Society 52: 57– 63 [Google Scholar]
- Thiers B.2011. [continuously updated]. Index Herbariorum: A global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium; http://sweetgum.nybg.org/ih/ [Google Scholar]
- Verkley GJM, Crous PW, Groenewald JZ, Braun U, Aptroot A.2004. Mycosphaerella punctiformis revisited: morphology, phylogeny, and epitypification of the type species of the genus Mycosphaerella (Dothideales, Ascomycota). Mycological Research 108: 1271– 1282 [DOI] [PubMed] [Google Scholar]
- Vilgalys R, Hester M.1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238– 4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW.1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (eds), PCR protocols: A guide to methods and applications. Academic Press, Inc., New York, New York: 315– 322 [Google Scholar]