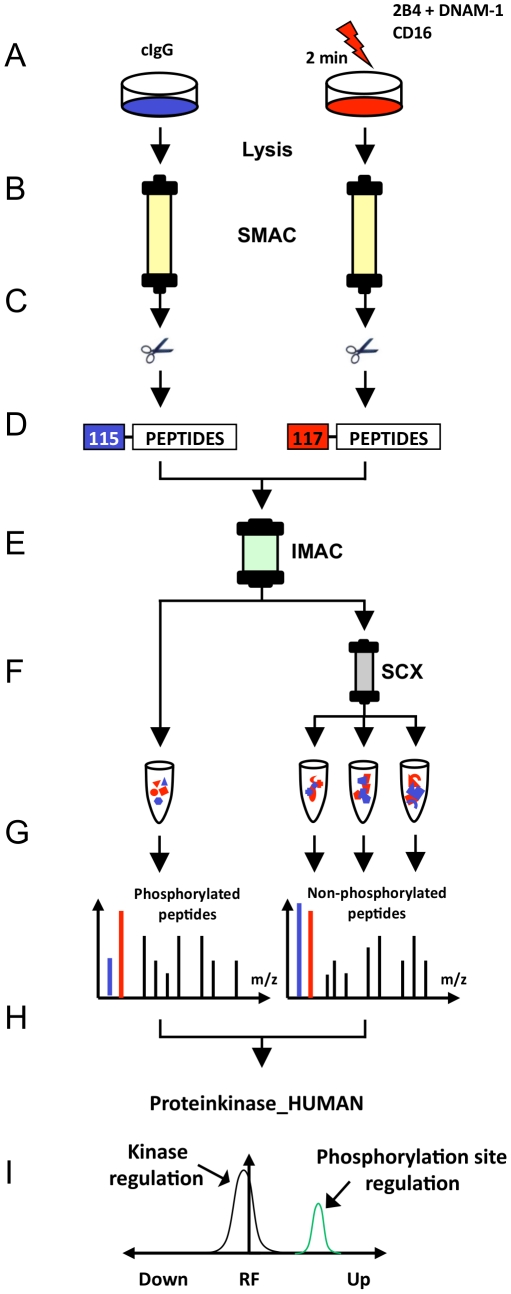
Figure 2. Proteome workflow for the detection of receptor-induced kinase phosphorylation in NK cells.
(A) Human, polyclonal, IL-2–cultured NK cells were stimulated each with the indicated receptor-specific monoclonal antibodies (mAbs) for 2 minutes (right side) and in all cases were comparatively analyzed with non-stimulated cells treated with control IgG isotype mAbs (cIgG) (left side). (B) Protein kinases were purified from total lysates using VI16743/Purvalanol-B-based small molecule affinity chromatography (SMAC). (C) Kinase elution and digestion into tryptic peptides comprising all non-modified and phosphorylated peptides. (D) Differential peptide labeling with iTRAQ reagents for MS-based quantification of phosphorylation site regulation and combination of samples. (E) Enrichment of phosphorylated peptides (phosphopeptides) by immobilized metal affinity chromatography (IMAC). (F) Further fractionation of non-phosphorylated peptide samples by strong cation exchange chromatography (SCX). (G) Peptide sequence analysis (LC-MS/MS) of all peptide fractions. (H) Peptide, kinase and phosphorylation site identification by Mascot database search and manual MS data inspection. (I) Statistical evaluation of significantly regulated phosphorylation sites on protein kinases by the MS-specific noise model iTRAQassist [38].