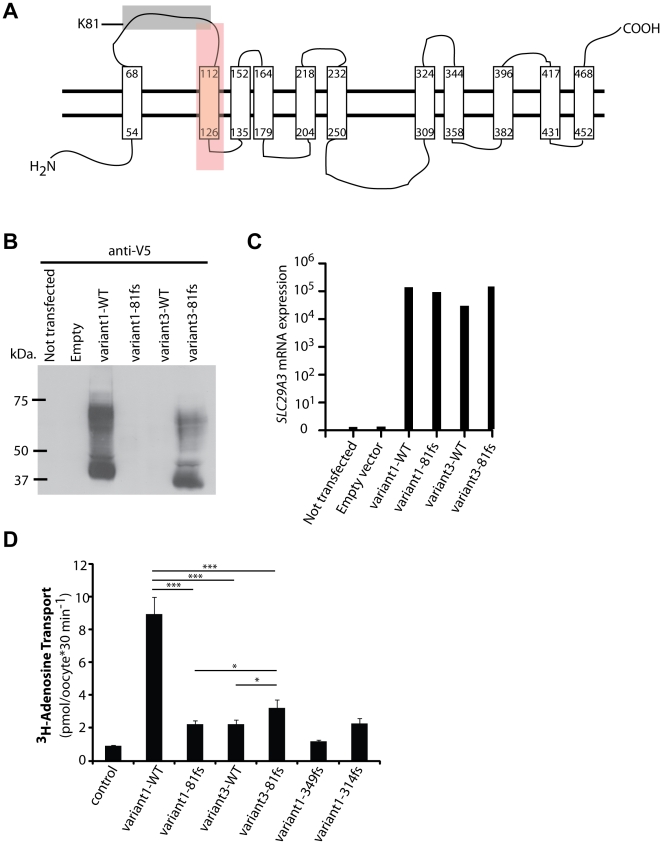
Figure 4. Expression and characteristics of hENT3-variant3-81fs.
(A) Scheme of hENT3-variant1-WT based on the protein structure predicted by SVMtm or TMpred programs. Transmembrane domains are represented by long rectangles. The gray and red boxes highlight the amino acids that are not identical in hENT3-variant1-WT and hENT3-variant3-81fs. In the gray box: amino acids 81–100 that are replaced by 20 other amino acids in hENT3-variant3-81fs. In the red box: amino acids 101–128 that are deleted in hENT3-variant3-81fs. (B) Levels of hENT3 proteins were assessed by immunoblotting with an anti-V5-tag antibody. The V5 tag was located at the C-terminus of hENT3. (C) The graph shows the relative SLC29A3 mRNA levels measured by Q-PCR for the same experiment with the V5-tagged constructs. Threshold cycles (Ct) for SLC29A3, normalized with respect to those of GUS (ΔCt), are plotted as 2−(ΔCt). Immunoblotting and Q-PCR results representative of three independent experiments are shown. (D) Adenosine transport activity. Uptake of [3H]adenosine (0.026 µM) in Xenopus oocytes 48 h after the injection of Δ36hENT3-variant1-WT or other variants with or without the 81fs mutation as well as one H (349fs) and one PHID syndrome (314fs) mutant cRNAs. pH-dependent uptake at pH 5.5. The data shown are the means ± SEM from three independent experiments. *, p<0.05, ***, p<0.001.