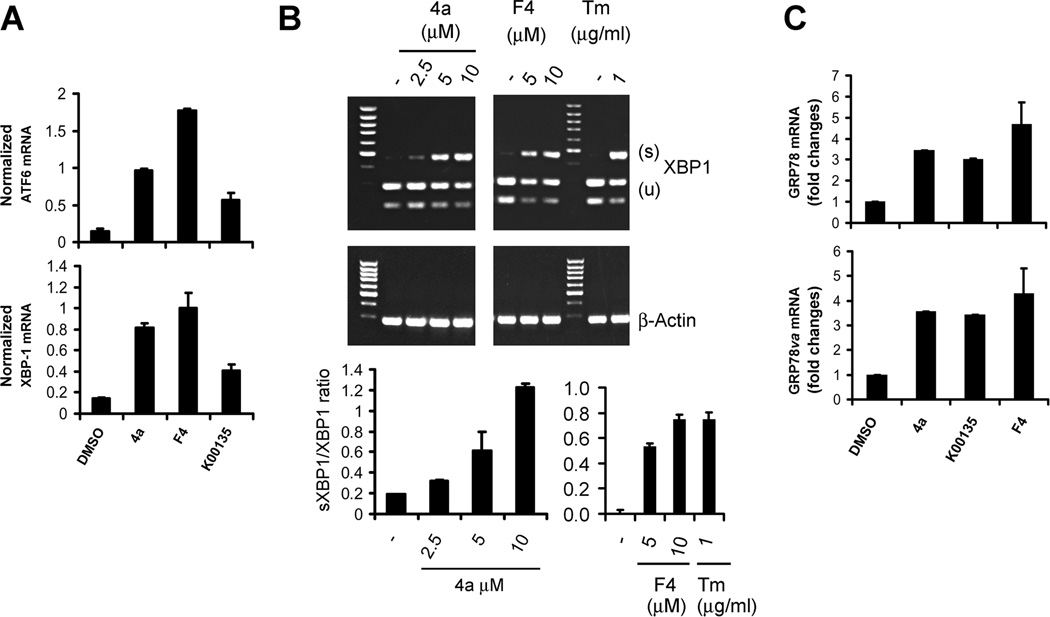
Fig. 5. Pim kinase inhibitors induce XBP-1 splicing.
A, LNCaP cells were treated with DMSO, 10 µM SMI-4a (4a), 10 µM K00135, and 10 µM 10058-F4 (F4) for 16 h. Total RNA was isolated and then subjected to qT-PCR. The levels of ATF6 or XBP-1 mRNA were normalized to the 18S mRNA. B, LNCaP cells were treated with SMI-4a (4a), 10058-F4 (F4) or Tunicamycin (TM) at the indicated doses for 16h. Total RNA was isolated and then subjected to RT-PCR for detection of XBP-1 splicing (top panel). The ratio of the spliced (S) to the unspliced (U) XBP-1 mRNAs was assessed by qT-PCR analysis (bottom panel). The mean of triplicate determinations are shown +/− standard deviation. C, Total RNA isolated in A was subjected to qT-PCR analysis to determine level of GRP78 and its isoform GRP78va using the 18S mRNA as an internal control.