Abstract
Purpose
EGFR targeted therapy is in clinical use to treat squamous cell carcinoma of the head and neck and other cancers of lining epithelium. Ras mutations in these tumors are a negative prognostic factor for response and skin inflammation is an adverse reaction to therapy. We investigated transcriptional and biochemical changes that could account for the confounding effects of RAS activation and inflammation in a squamous tissue.
Experimental Design
We performed gene expression profiling on oncogenic Ras transformed and wildtype mouse and human keratinocytes with EGFR ablated chronically by genetic deletion or acutely by drug treatment and followed leads provided by pathway analysis with biochemical studies.
Results
We identified a 25 gene signature specific to the Ras-EGFR ablation interaction and a distinct 19 gene EGFR ablation signature on normal keratinocytes. EGFR ablation in the context of wildtype Ras reduces ontologies favoring cell cycle control and transcription while oncogenic Ras enriches ontologies for ion channels and membrane transporters, particularly focused on calcium homeostasis. Ontologies between chronic EGFR ablation and acute pharmacological ablation were unique, both with and without Ras activation. p38α is activated in response to abrogation of EGFR signaling under conditions of Ras activation in both mouse and human keratinocytes and in RAS transformed tumor orthografts of EGFR ablated mouse keratinocytes. EGFR ablation in the absence of oncogenic Ras revealed Erk and IL-1β related pathways.
Conclusion
These findings reveal unrecognized interactions between Ras and EGFR signaling in squamous tumor cells that could influence the therapeutic response to EGFR ablation therapy.
Keywords: EGFR, Ras, keratinocytes, gene profiling
Introduction
The epidermal growth factor receptor (EGFR) is a transmembrane cell surface tyrosine kinase in the ErbB family. At least 5 ligands are known to activate EGFR kinase activity directly, and kinase function can be activated indirectly through interactions with neighboring receptors. Upon ligand binding, EGFR may either homo- or hetero-dimerize resulting in autophosphorylation on several specific tyrosine residues. A number of intracellular substrates are phosphorylated by activated EGFR kinase resulting in stimulation of downstream pathways, including the RAS/MAPK pathway and PI(3)K/AKT pathway among others (1). EGFR activity is increased in a number of human cancers by amplification, mutation or ligand excess, and therefore EGFR has become a target for anticancer therapy. Erlotinib (Tarceva, OSI-774), currently approved to treat non-small-cell lung cancer (NSCLC) and advanced pancreatic cancer (2), selectively and reversibly inhibits the EGFR tyrosine kinase by binding the intracellular ATP binding domain (3). Cetuximab, an anti-EGFR monoclonal antibody that blocks ligand-induced EGFR activation, is currently approved for metastatic colorectal cancer (mCRC) and head and neck squamous cell carcinoma (HNSCC). RAS mutations occur in 15–30% of NSCLC (4), in 30–50% of mCRC (5), 60–80% of pancreatic cancers (6, 7) and 5–8% of HNSCC. In NSCLC, HNSCC and mCRC RAS mutations are negative prognostic factors (8). Patients with tumors harboring RAS mutations that are treated with EGFR targeted therapy are less or non-responsive or may have decreased survival as compared to chemotherapy alone (9) (10). In addition, patients on anti-EGFR therapy often suffer from a severe acneiform, papulopustular folliculitis that involves the face, scalp, chest, back and arms, characterized by neutrophilic infiltration in the infundibular portion of the hair follicle, alopecia and xerosis cutis unresponsive to standard therapy (11, 12). While the extent of the skin rash often correlates with positive clinical response in responsive patients (13), evaluating the RAS gene status before dispensing an EGFR inhibitory drug will reduce rash-associated morbidity.
A promising model to evaluate the interaction of the RAS and EGFR pathways experimentally is the EGFR null mouse that recapitulates the dose-limiting rash of anti-EGFR therapy as a fulminant papulopustular neutrophilic folliculitis that consumes the hair follicles leading to alopecia (14). Oncogenic ras transformed keratinocytes from EGFR null mice produce squamous tumors as orthografts, and these tumors have a high proliferation rate in the absence of EGFR even though the tumors are very small (15). We hypothesized that specific changes in gene expression induced by oncogenic ras in this squamous tumor model could illuminate pathways contributing to the poor clinical response of tumors harboring RAS mutations in patients on anti-EGFR therapy. In the current study, we have compared the cell autonomous phenotype of EGFR genetic deficiency with acute pharmacological ablation of the pathway by anti-EGFR therapeutics in skin keratinocytes with and without an activated mutant Ras oncogene. We find substantial transcriptional differences among these groups and define a signature for each. Further, pathway analysis and biochemical experiments revealed an unexpected activation of p38 signaling that is unique to the oncogenic ras-EGFR ablated phenotype. Suppression of p38 signaling inhibits growth of EGFR ablated keratinocytes oncogenically transformed by activated Ras.
Materials and Methods
Materials
Erlotinib (Tarceva) was supplied by OSI Pharmaceuticals Inc (Melville, NY), AG1478 was purchased from Sigma-Aldrich (St Louis, MO), PD153035 and SB203580 were purchased from Calbiochem Inc. (San Diego, CA). All drugs were dissolved in DMSO.
Mouse Model
Mouse studies were performed under a protocol approved by the National Cancer Institute (NCI) and NIH Animal Care and Use Committee. Homozygous and heterozygous EGFR null mice and wild-type siblings on CD-1 background have been described previously (14).
Cell Culture
Primary mouse keratinocytes were isolated from newborn EGFR-deficient and wild-type mice. Keratinocytes were prepared as described previously (16) and cultured in calcium- and magnesium- free MEM (Invitrogen Life Technologies, Carlsbad, CA) supplemented with 8% Chelex (Bio-Rad Laboratories, Hercules, CA)-treated FBS (Gemini Bioproducts) and 0.2 mM Ca2+. After 24 h, cultures were switched to the same medium with 0.05 mM Ca2+ to select for basal cells and cultured for 2 additional days. Transduction of v-rasHa into keratinocytes (termed ras-keratinocytes) employed a replication defective retrovirus as described previously (17). Viral infections were performed using diluted supernatant from Ψ-2 producer cells. Cells were cultured for 4 days following v-rasHa transduction and then treated with 1 µM of the various EGFR tyrosine kinase inhibitors for 2 hours or 14 hours. Non-transduced cells were treated on day 4 after plating and at the same concentration and time points as the v-rasHa-transduced keratinocytes. The same protocol was used to infect cells with retrovirus encoding K-rasv12 generated by transfecting a Platinum A retroviral packaging cell line (Cell Biolabs, Inc) with pBabe K-Ras V12 (Addgene) and harvesting medium 48 hours later for infection of keratinocytes. Human keratinocyte HaCaT cells (18) were cultured in DMEM with 10% FBS. Human RAS transformed keratinocytes HaCaT-II-4 were provided by Dr. Jonathan Garlick (Tufts University) and cultured in low-glucose DMEM with 5% FBS, 10mM HEPES, and Pen/strep (Life Technologies/Invitrogen).
Microarray Design and Biometric Analysis
Ten µg total RNA from tissue culture samples or mouse universal reference total RNA (Stratagene, La Jolla, CA) were reverse transcribed and the cDNA coupled to Cy3 or Cy5 dyes (Amersham Biosciences, Picataway, NJ) respectively, using the indirect labeling procedure as previously described (19). The labeled Cy3 and Cy5 cDNA was purified using QIAGEN PCR Cleanup kit (QIAGEN Inc. Valencia, CA). For each microarray, Cy3 labeled cDNA was co-hybridized with Cy5 labeled cDNA and compared on each slide. Reverse fluor experiments were included for each group. Each group contained 5 biological repeats. The labeled cDNA’s were mixed with Mouse Cot-1 DNA, yeast tRNA and polydA and boiled for 2 minutes prior to hybridization on microarray slides at 42°C for 14–16 h. Poly-L-lysine coated glass slides, containing approximately 37K mouse oligoarray features, were obtained from the NCI Microarray Core Facility. After hybridization, the slides were washed in SSC, dried, and scanned using GenePix 4000 microarray scanner (Axon Instruments, Union City, CA) with Genepix 4.0 software. Spots with a size of <10 µm or an intensity of <100 in both the red and green channels were eliminated, followed by removal of features that were uninformative in >50% of available arrays. Each array was normalized using a Lowess with print-tip group to provide intensity-dependent normalization.
Statistical analyses were done using the BRB-ArrayTools package (version 3.4.0) developed by Dr. Richard Simon and BRB-ArrayTools Development Team for microarray analysis. Class comparison analysis was done using the expression data. The two-sample t statistic with random variance model (20) was used to measure the difference in gene expression between two classes. Genes were considered as being differentially expressed at a significance of P ≤ 0.01.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (21) and are accessible through GEO Series accession number GSE29415.
Ontology
GoMiner and EASE analyses were done on the lists of differentially expressed genes (22, 23). Enrichment of categories was determined using GoMiner, and P value thresholds were determined from EASE scores.
Ingenuity Pathway Analysis
Data sets containing the NCI probe set identifiers and fold changes were uploaded into Ingenuity Pathway Analysis software (IPA). The IPA program searches the Ingenuity Pathway Knowledge Base (IPKB) for interactions between the uploaded genes and all other genes in the IPKB and generates a series of networks. Fisher’s exact t-test was used to assign statistical significance and biological functions were also assigned to each network.
Quantitative real-time PCR
RNA was isolated using Trizol (Invitrogen). cDNA synthesis and real time PCR analysis was previously described (24).
Immunoblotting
Cultures were washed twice with PBS, and total cell lysates prepared with 1% triton containing lysis buffer (Cell Signaling) supplemented with 1mM PMSF, Halt Phosphatase Inhibitor (Pierce) and Mini-complete tablets (Roche). Proteins were quantified with the Bradford method (Bio-Rad) and separated by a 4–20% SDS-PAGE gel. Phospho-EGFR (Invitrogen), EGFR, p38α, p38δ (Santa Cruz), p38 and phospho-p38 (Cell Signaling) were used at 1:1,000 overnight at 4°C, β-actin (Chemicon) was used 1:10,000 for one hour room temperature. ECL SuperSignal (Pierce) system was used for detection.
Immunohistochemistry
Serial sections of five micrometer paraffin embedded tumors were treated with 3% hydrogen peroxide to inhibit peroxidase activity, blocked in Dako blocking solution (Dako) and incubated overnight with phospho-p38 (Cell Signaling) and phospho-EGFR antibodies. The next day sections were treated with goat anti-rabbit-HRP and sheep anti-mouse HRP (GE Healthcare) respectively at 1:500 dilution. Immunoreactivity was detected using the DAB kit (Vector Laboratory).
3H-Thymidine incorporation assay
Keratinocytes, EGFR wildtype and EGFR null, were plated in 24 well plates and transduced with v-rasHa or K-rasV12 as described. Five days later cells were treated with the appropriate EGFR TKI for 14hrs. For co-treatments cells were pre-treated with p38 inhibitor for 4 hours at 10µM. 3H-thymidine (1 µCi per well) was added for 3hrs. Cells were fixed using methanol and acetic acid (3:1 ratio), solubilized with NaOH (5 normal), and incorporated counts were read using a scintillation counter.
siRNA
p38α siRNA 5’-CTCCAGCTACTTTGTGTTGAA-3’, p38β siRNA 5’-CAGATTATCTATCTAGTCAGA-3’, p38δ siRNA 5’-CTGGCTAGACCTGCAACTCAA-3’, and All Stars negative control siRNA were obtained from Qiagen and transfected using HiPerfect (10nM; Qiagen) according to the manufacturers protocol. All siRNA transfections were performed three days after v-rasHa transduction. 32hrs post-transfection, EGFR wildtype cells were treated with AG1478, Erlotinib, or PD153035 for 14hrs, rinsed with PBS and collected for total protein isolation.
Results
Gene expression profiles of EGFR ablated ras transformed keratinocytes
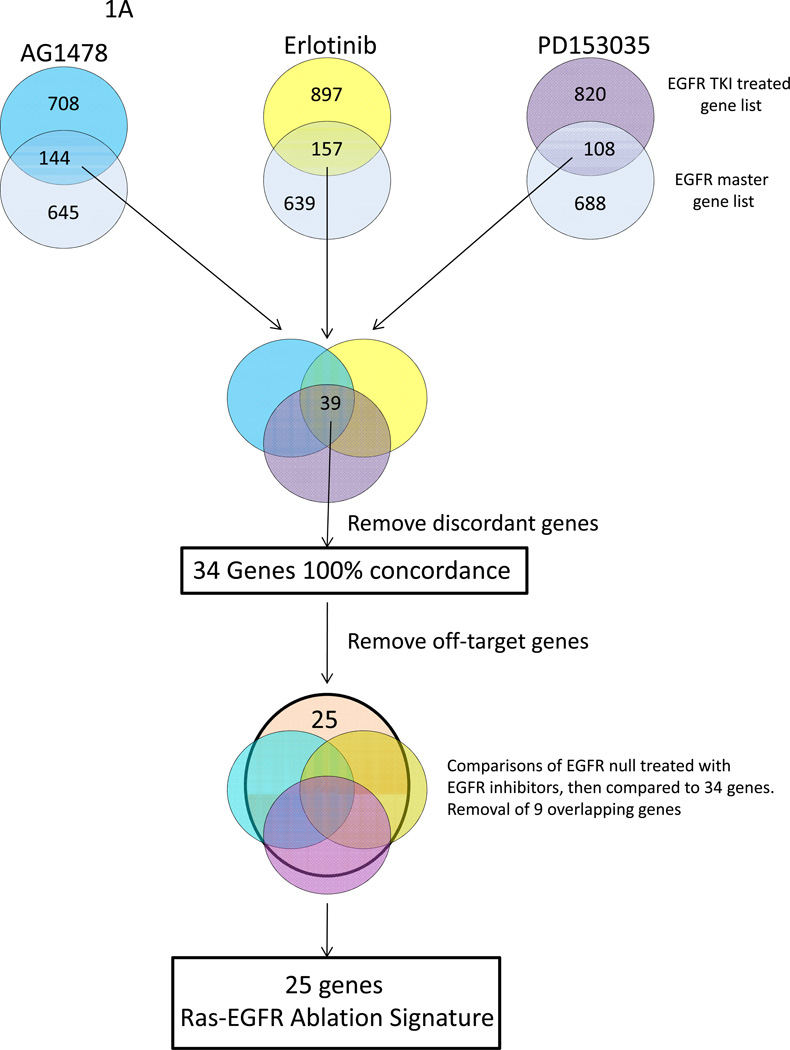
Our goal was to identify a gene expression signature common to genetic and pharmacologic ablation of EGFR in the context of oncogenic Ras transformation. To achieve this end, we transformed wildtype mouse keratinocytes or EGFR null keratinocytes with a v-rasHa retroviral vector and isolated RNA 2 hours after treatment with EGFR inhibitors. To identify differentially expressed genes, class comparisons were performed and genes selected with a p ≤ 0.01. When compared to EGFR wildtype keratinocytes transduced with v-rasHa, this analysis yielded differentially expressed gene lists for AG1478, Erlotinib, PD153035 and EGFR null of 852, 1054, 928, and 758 respectively (Figure 1A). To determine gene expression off-target effects of the EGFR pharmacological inhibitors, EGFR null v-rasHa keratinocytes were treated with AG1478, Erlotinib or PD153035. Class comparisons between treated and untreated EGFR null v-rasHa keratinocytes gave off-target gene lists of 891, 380 and 2,486 of differentially expressed genes for AG1478, Erlotinib and PD153035 respectively.
Figure 1.
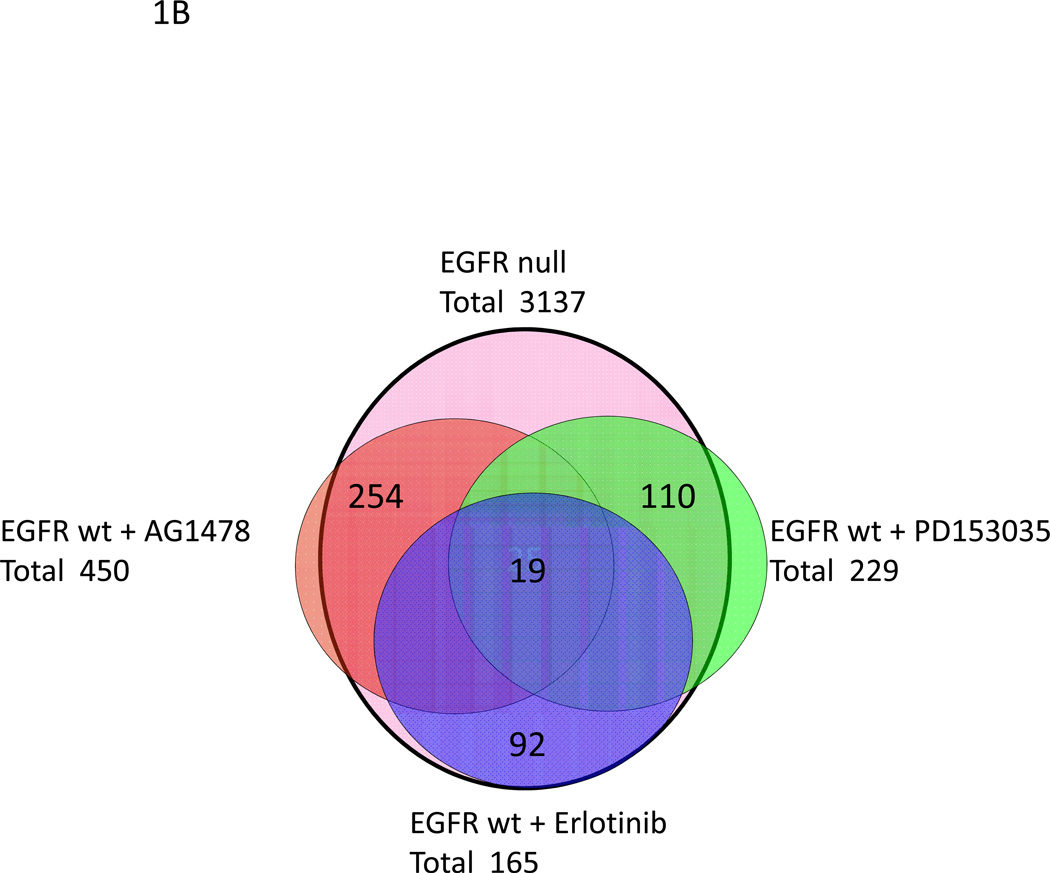
Analytic scheme used to identify common gene expression changes in oncogenic Ras transformed (A) or wildtype keratinocytes (B) lacking EGFR signaling by genetic ablation or pharmacological inhibition. A, v-rasHA transformed keratinocytes were treated with each EGFR inhibitor for 2 hours, RNA was isolated and expression changes relative to untreated transformed keratinocytes were documented using the 37K mouse oligo array platform. A master EGFR gene list was similarly generated from the profile of v-rasHa transformed EGFR null keratinocytes that overlapped a comprehensive map of EGFR signaling (27) (light blue), and these lists were intersected to identify common genes that were EGFR relevant. These three overlapped gene lists were then intersected, shown in the Venn diagram, discordant genes were removed and a resultant list of 34 common genes were identified. By treating oncogenic Ras transformed, EGFR null keratinocytes with the three EGFR inhibitors, off-target gene expression changes were identified, and 9 genes in the 34 concordant gene list were removed as off target genes. This resulted in 25 genes that comprised the Ras-EGFR ablation signature. B, a Venn diagram illustrates the similar analysis used to determine the common and concordant genes between the four methods of EGFR ablation in the absence of oncogenic Ras. Each gene expression profile is from five biological repeats.
To understand the activated Ras effect on keratinocytes, it was also necessary to identify a signature common to genetic and pharmacologic ablation of EGFR without Ras-mediated transformation. To achieve this, four days after plating we isolated RNA 2 hours after treatment of wildtype keratinocytes with EGFR inhibitors. RNA was also collected from EGFR null keratinocytes on day four after plating. As in the Ras transformed comparisons, all differentially expressed gene class comparisons were performed with a p≤0.01. When compared to untreated EGFR wildtype keratinocytes, treated keratinocytes yielded differentially expressed gene lists for AG1478, Erlotinib, PD153035, and EGFR null of 450, 165, 229, and 3,137 respectively (Figure 1B). These array comparisons express the difference between an acute blockade of EGFR by inhibitors and a chronic shutdown of the EGFR signaling by genetic ablation in keratinocytes. Whereas the EGFR inhibitor treated groups have relatively small numbers of gene expression changes, the EGFR null array comparison has over ten-fold higher number of changes, expressing the myriad of consequences of chronic inhibition of EGFR on wildtype keratinocytes. Further, the introduction of oncogenic ras into the genome adds significantly more complexity to the acute response to inhibitors but reduces the number of differentially expressed genes in keratinocytes chronically depleted of EGFR by gene ablation. This suggests that there are a significant number of common changes in gene expression produced by Ras transformation and EGFR activation.
To verify our microarray data, we conducted quantitative real-time PCR for 25 differentially expressed genes. Supplemental Table 1 shows the fold change for both the real time PCR and the microarray. All genes screened were concordant in direction of expression and for the most part similar in magnitude between the real time data and microarray data.
Identification of signatures for EGFR ablation
To identify commonly expressed genes between the different methods for EGFR ablation, an EGFR master list was created. The master list was created by combining the differentially expressed genes of the microarray comparison between EGFR wildtype v-rasHa keratinocytes and EGFR null v-rasHa keratinocytes, and genes that have been cataloged from a comprehensive map of EGFR signaling (25). This EGFR master list of 789 genes serves as the basis for further analysis and comparison with the samples treated with chemical EGFR inhibitors. A four step analysis (Figure 1A) was performed; comparison of gene lists from AG1478, Erlotinib or PD153035 treated v-rasHa keratinocytes with EGFR master list, intersecting the overlapping genes between the EGFR inhibitors and the EGFR master list with each other to find commonalities, and removal of discordant genes. At this point the analysis had yielded 34 genes. To ensure no genes involved in off-target effects were included, we intersected the 34 genes with the three off target gene lists; this filtered out 9 genes to achieve a 25 gene list. This gene list is a signature for EGFR ablation in keratinocytes when v-rasHa is activated (Table 1A).
Table 1.
Signature gene lists for keratinocytes inhibited for EGFR signaling and expressing either oncogenic Ras (A) or wildtype Ras (B). Downregulated genes are in green and upregulated in red. Values are relative to EGFR wild-type keratinocytes with oncogenic (A) or wild-type Ras (B).
| Table 1A. | ||||
|---|---|---|---|---|
| Gene | EGFRnull | Erlotinib | AG1478 | PD153035 |
| Downregulated | ||||
| dual specificity phosphatase 1 (Dusp1) | −3.34999 | −2.113999 | −2.409 | −2.659 |
| thioredoxin domain containing 10 (Txndc10) | −3.208999 | −2.964 | −4.531 | −2.412999 |
| PREDICTED: neuregulin 1, transcript variant 2 (Nrg1) | −3.187999 | −2.862 | −2.984999 | −2.765999 |
| PREDICTED: E3 ubiquitin protein ligase, HECT domain containing, 1, transcript variant 6 (Edd1) | −1.807 | −2.83999 | −2.264 | −1.831 |
| Up Regulated | ||||
| solute carrier family 30 (zinc transporter), member 1 (Slc30a1) | 2.2675737 | 2.2522523 | 2.8248588 | 1.984127 |
| surfeit gene 1 (Surf1) | 2.2883295 | 2.1786492 | 1.8382353 | 2.1008403 |
| Ubiquitin domain containing 1 | 2.3696682 | 2.6315789 | 3.5971223 | 2.5316455 |
| inhibitor of DNA binding 2 (Id2) | 2.7027027 | 4.950495 | 4.2553191 | 3.8167939 |
| PREDICTED: RIKEN cDNA 4930458A03 gene (4930458A03Rik), mRNA. | 2.8490028 | 3.076923 | 2.4752475 | 4.2194092 |
| aldehyde oxidase 1 (Aox1) | 2.7322404 | 2.9411765 | 2.7548209 | 2.5906736 |
| RIKEN cDNA 2200002J24 gene (2200002J24Rik), mRNA. | 2.8735632 | 3.4013605 | 3.78787878 | 2.3364485 |
| RIKEN cDNA 6230410P16 gene (6230410P16Rik), transcript variant 2 | 2.8985507 | 2.7624309 | 2.5974026 | 2.7173913 |
| fucosyltransferase 9 (Fut9) | 3.030303 | 3.8910506 | 2.0746888 | 3.0674847 |
| Tissue inhibitor of metalloproteinase 3 | 3.2679739 | 3.2051282 | 3.2258065 | 2.3419204 |
| RIKEN cDNA 4921528I07 gene | 3.2786885 | 3.5460992 | 3.1545741 | 3.7735849 |
| synaptoporin (Synpr) | 3.3670034 | 2.8011204 | 3.3444816 | 3.030303 |
| testis specific gene A2 (Tsga2) | 3.3670034 | 4.4247788 | 3.6630037 | 4.4247788 |
| ATPase, Ca++ transporting, plasma membrane 3 (Atp2b3) | 3.5335689 | 3.4482759 | 4.0650407 | 3.968254 |
| interferon-induced protein 44 (Ifi44) | 3.5335689 | 3.5714286 | 3.4246575 | 2.6041667 |
| CD46 antigen, complement regulatory protein (Cd46) | 3.6101083 | 4.3668122 | 3.8314176 | 3.90625 |
| RIKEN cDNA 2310026E23 gene (2310026E23Rik), mRNA. | 4.2016806 | 3.5842293 | 3.3898305 | 2.1881838 |
| solute carrier family 16 (monocarboxylic acid transporters), member 12 (Slc16a12), mRNA. | 5.1282051 | 5.0761421 | 6.2789447 | 3.703703 |
| camello-like 1 (Cml1) | 5.5555556 | 5.3475936 | 5.9171598 | 13.69863 |
| ryanodine receptor 1, skeletal muscle (Ryr1) | 3.7522278 | 3.2258065 | 3.6764706 | 3.4722222 |
| F-box and leucine-rich repeat protein 13 (Fbxl13), mRNA. | 11.6279069 | 5.1020408 | 10.2040816 | 5.4644808 |
| Table 1B. | ||||
|---|---|---|---|---|
| Gene | EGFR null | Erlotinib | AG1478 | PD153035 |
| Downregulated | ||||
| tenascin C (Tnc), mRNA. | −41.87473 | −9.9395794 | −9.0436273 | −6 7006022 |
| chemokine (C-X-C motif) ligand 3 (Cxcl3), mRNA. | −12.4585 | −6.0032651 | −7.2140103 | −5.4960882 |
| serine (or cysteine) peptidase inhibitor, clade E, member 1 (Serpine1), mRNA. | −5.480587 | −3.9510035 | −4.261114 | −3.6979597 |
| dual specificity phosphatase 9 (Dusp9), mRNA. | −7.642714 | −3.7126055 | −6.4856947 | −4.8857617 |
| forkhead box D3 (Foxd3), mRNA. | −3.025117 | −2.6639708 | −2.8218732 | −2.624347 |
| Golgi integral membrane protein 4 | −6.650365 | −3.2812439 | −3.4076097 | −3.0076131 |
| cell division cycle 6 homolog (S. cerevisiae) (Cdc6), transcript variant 2, mRNA. | −3.177363 | −2.9617525 | −3.3367854 | −2.6737621 |
| kinesin family member 11 (Kif11), mRNA. | −2.586524 | −2.3925532 | −2.801853 | −2.1541163 |
| Upregulated | ||||
| BEN domain containing 7 (Bend7), mRNA. | 2.986081 | 2.59399268 | 2.17177856 | 2.123838473 |
| Transcribed locus | 5.117288 | 2.62182163 | 2.62978239 | 2.357328025 |
| RRN3 RNA polymerase I transcription factor homolog (yeast) (Rrn3), mRNA | 4.179157 | 3.01058099 | 3.03574806 | 2.600699744 |
| membrane-spanning 4-domains, subfamily A, member 13 (Ms4a13), mRNA. | 10.55176 | 2.62303544 | 5.03417703 | 2.540895071 |
| adenosine A3 receptor (Adora3), transcript variant 2, mRNA. | 6.572833 | 3.02683959 | 3.77621258 | 3.789070049 |
| Vacuolar protein sorting 13A (yeast) | 2.670708 | 2.63874731 | 2.15684629 | 1.896646066 |
| adrenergic receptor, alpha 2b (Adra2b), mRNA. | 3.285338 | 3.01698624 | 3.86207003 | 2.693184117 |
| LY6/PLAUR domain containing 6 (Lypd6), mRNA. | 2.30441 | 2.1395848 | 2.13043367 | 2.320750995 |
| engrailed 2 (En2), mRNA. | 2.444314 | 2.30637466 | 2.00310682 | 2.242360614 |
| somatostatin (Sst), mRNA. | 3.569832 | 3.90594027 | 3.58261785 | 2.805393762 |
| myosin light chain kinase 3 (Mylk3), mRNA. | 1.791513 | 2.14708521 | 1.92247208 | 3.027817467 |
To create a signature for EGFR ablation in the absence of oncogenic ras, we performed similar comparisons as above. The array comparison between EGFR wildtype keratinocytes and EGFR null keratinocytes yielded 3,137 differentially regulated genes, and there was significant overlap with the comprehensive map (25). The three EGFR inhibitor array gene lists were intersected with the EGFR null gene list, resulting in an overlap of 19 genes (Figure 1B). All 19 genes were concordantly expressed between the different methods of EGFR ablation (Table 1B). Surprisingly there was no overlap among the gene lists with and without oncogenic Ras although both showed down-regulation of a member of the dual specificity kinase family (Dusp 1 and Dusp 9). This suggests that oncogenic Ras exerts a strong influence on the consequences of EGFR inhibition in keratinocytes.
Gene ontology and pathway analysis
Gene ontology categories from the total gene lists of Figure 1A and B that are significantly enriched in the four methods of EGFR ablation in the presence of oncogenic Ras are shown in Supplemental Table 2A, 2B, 2C, and 2D. The cut-off for all ontology analysis was an EASE score ≤ 0.05. The color is based on the average fold change of the genes matching to specific ontologies, thereby giving information as to the over-all expression direction of the ontology. Each inhibitor had a unique ontology profile although some general overlapping areas could be discerned. Among the gene categories for EGFR ablation are ATPase activity, regulation of transcription, MAPKKK cascade, cell proliferation, ion transport and enzyme inhibitor activity. To better understand the effects of pharmacological inhibition, we catalogued the genes that are common and concordant between the three inhibitors. Supplemental Table 2E shows the ontologies enriched from the 109 genes that overlapped the three methods of acute pharmacological blockade. A predominant theme arising from this analysis is the frequency of enrichment in ontologies for ion, cation, and chemical homeostasis. To further understand the core differences between acute (Erlotinib) and chronic (EGFR null) inhibition in the presence of oncogenic Ras, we removed overlapping or related ontologies and showed the remaining unique ontologies in Supplemental Table 2F. Chronic depletion by gene ablation depresses genes encoding proteins involved in transcription, cell growth and cell cycle regulation. In contrast, acute exposure to Erlotinib emphasizes an up-regulation of genes encoding metabolic enzymes and stress responses, a change common to all of the inhibitors.
Supplemental Tables 3A, 3B, 3C, and 3D are enriched ontologies from the four methods of EGFR ablation in the absence of oncogenic Ras transformation. There is almost no overlap in enriched ontologies between the different methods of EGFR ablation. Only a few gene ontologies, particularly related to cell adhesion and motility, are commonly enriched. The three chemical inhibitors do share similar immune related ontologies, which are chemokine activity, inflammatory response, and immune response. These changes could be associated with an inflammatory response mediated through EGFR inhibition in skin keratinocytes. In the absence of oncogenic Ras transformation, the diversity and extent of gene ontology changes is much greater when comparing EGFR wildtype to null keratinocytes than when comparing EGFR wildtype keratinocytes to keratinocytes acutely ablated for EGFR. By examining ontologies based only on the 25 genes v-rasHa-EGFR ablation signature (Supplemental Table 4), five enriched ontologies encompassing the same genes becomes apparent. These ontologies define common functions as membrane transporters.
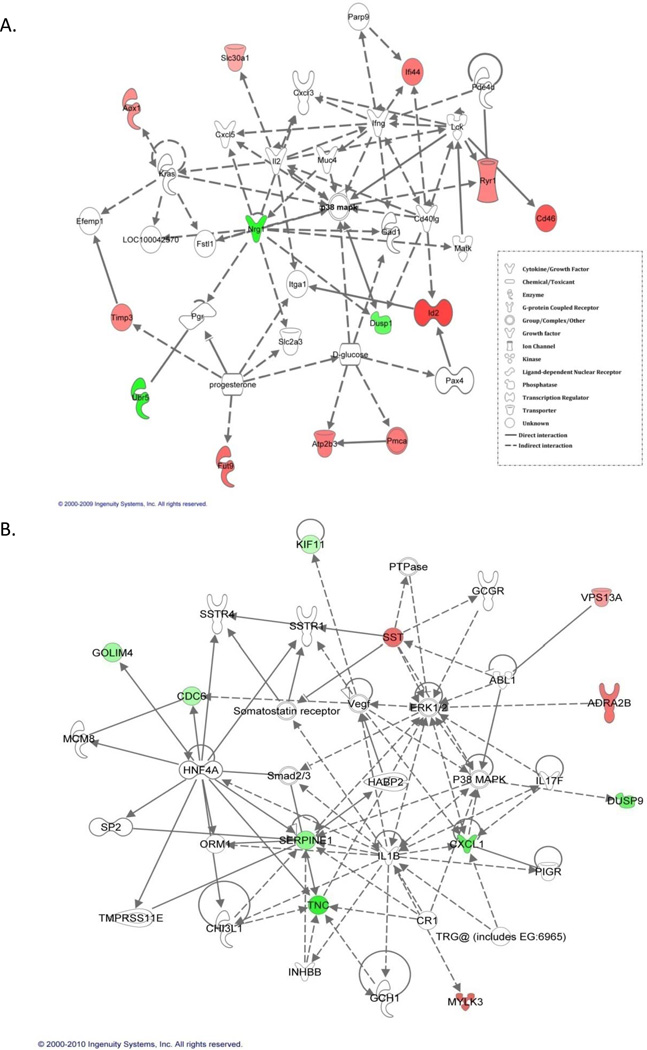
Using Ingenuity Pathway Analysis (IPA) and its Ingenuity Pathway Knowledge Base (IPKB) we investigated interactions among the v-rasHa- EGFR ablation signature gene list and found that 14 of the 25 genes mapped to one network shown in Figure 2A. With a score of 33, this network relates to cancer, cellular growth and proliferation, cellular development and tumor morphology. Central to this network is the p38 MAPK. The same analysis was performed on the EGFR ablation signature gene list in the absence of v-rasHa transformation and illustrated that 11 of the 19 genes mapped to one network. The network (Figure 2B) was the most significant and has a score of 26. Top functions of this network related to immune cell trafficking, cellular growth and proliferation, and cellular assembly and organization. Central to this network are Erk1/2 and IL-1β.
Figure 2.
Ingenuity pathway analysis (IPA) of the EGFR ablation signature with oncogenic (A) and wildtype Ras (B). Nodes represent genes, with shapes showing IPA-defined functional class of genes, and lines indicating biological relationships between nodes. A, a network based on the Ingenuity Pathway Knowledge Base of 14 of the 25 (56%) genes from oncogenic Ras-EGFR ablation signature. B, a network comprised of 11 of the 19 (57%) genes from the EGFR ablation signature with wildtype Ras.
p38MAPK is activated by EGFR ablation in the presence of ras activation
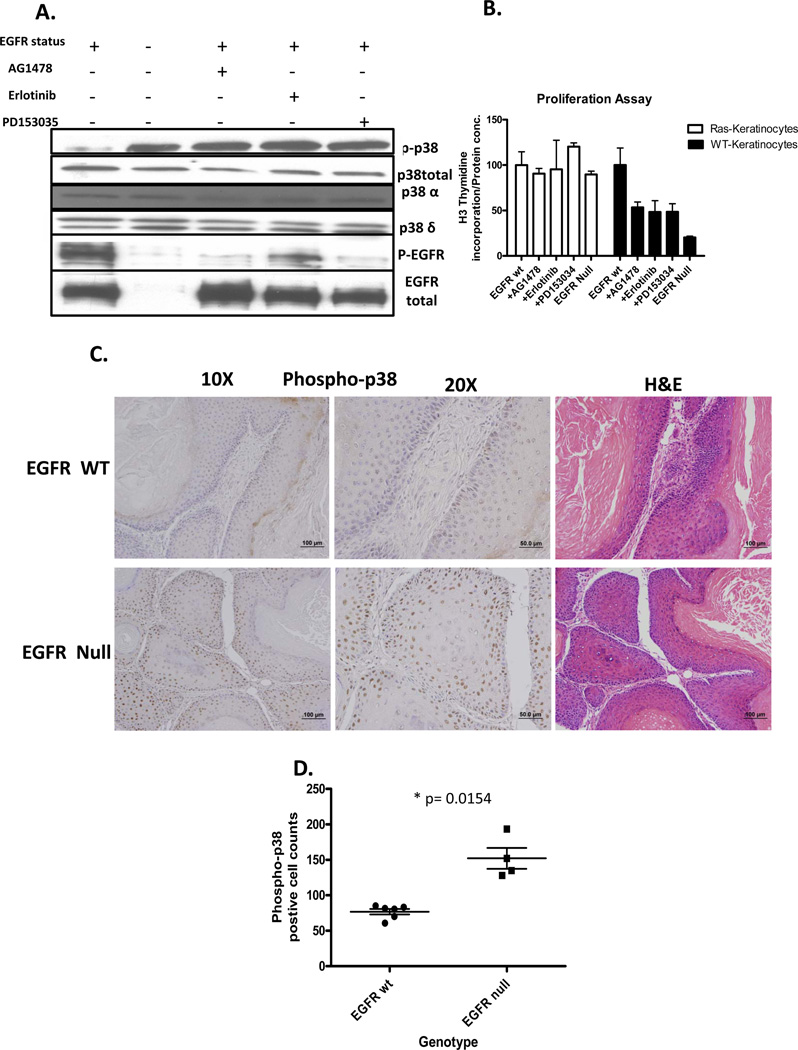
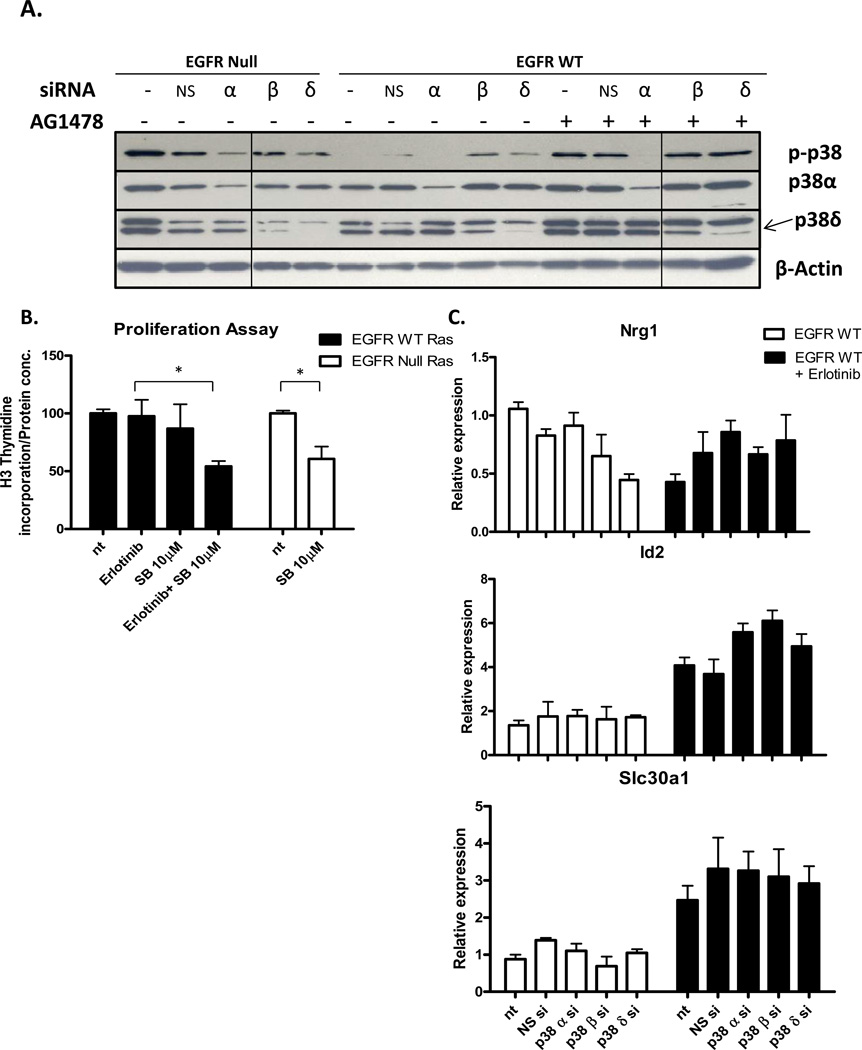
The MAPK p38 is a potent regulator of proliferation, inflammation, cell motility and differentiation. The p38 MAPK family includes p38α, β, δ, and γ, with three isoforms expressed in keratinocytes, α, β, and δ. The isoforms affect different cellular processes. Based on IPA mapping of p38 interaction with the v-rasHa- EGFR ablation signature, we tested the effects of EGFR inhibition on modification of the p38 isoforms. Pharmacological or genetic ablation of EGFR in combination with v-rasHa increases phosphorylation of p38 in cultured primary keratinocytes without changes in overall levels of p38α, p38β or p38δ (Figure 3A). Proliferation as determined by 3H-thymidine incorporation (Figure 3B) did not change in Ras transformed keratinocytes with either genetic or pharmacological ablation of EGFR. In contrast, genetic and pharmacological ablation of EGFR inhibited proliferation in wildtype keratinocytes. To confirm that the changes in p38 activation also occurs in vivo, squamous tumors arising from orthotopic grafts of EGFR wild type and null v-rasHa - transformed keratinocytes were immunostained for phospho-p38 (Figure 3C, D). In the absence of EGFR, phospho-p38 is abundantly detected in basal and suprabasal compartments of v-rasHa- oncogene induced tumors whereas detection is limited to a smaller number of cells restricted to the basal cell compartment in v-rasHa- induced tumors expressing EGFR. Consistent with the in vitro results, previous analysis of these tumors indicated that proliferation rate in tumor epithelium was independent of EGFR status (15).
Figure 3.
Inhibition of EGFR in the presence of oncogenic Ras activates p38. A, Immunoblots for total and phosphorylated p38 isoforms in keratinocytes inhibited for EGFR signaling. B, Proliferation of cultured wildtype or v-rasHa - transduced EGFR null or wildtype keratinocytes 24hr after treatment with EGFR pharmacologic inhibitors. C, Immunostaining for phospho-p38 in squamous tumors from orthotopic grafts of EGFR wild-type and EGFR null v-rasHa – transformed keratinocytes. D, Phospho-p38 labeled cells were counted in nine to ten randomly selected regions. Each group contains four (EGFR null) and six (EGFR wt) tumors, and data are representative of two independent grafting experiments. Horizontal lines= mean, vertical bars= SE.
p38α is the activated isoform of p38 in EGFR ablated cells
To determine the specific p38 isoform activated by oncogenic Ras when EGFR signaling is abrogated, isoform specific p38 siRNA was used to selectively silence transcription. In either EGFR null cells or EGFR wildtype cells transformed with v-rasHa, the level of phospho-p38 was reduced by siRNA targeting p38α but not p38β or p38δ (Figure 4A). Since p38β is undetectable by immunoblot we confirmed the effectiveness and specificity of the siRNA by real time PCR (Supplemental Figure 1). We inhibited p38 pharmacologically in v-rasHa transformed keratinocytes to determine if combination treatment with EGFR ablation would lead to growth inhibition. Blockade of EGFR or p38 alone did not reduce proliferation, but combining inhibition of EGFR and p38 reduced proliferation by 40–45% (Figure 4B). We also asked if p38 isoform knock-down would alter a sampling of genes differentially expressed as the EGFR ablation signature in wildtype keratinocytes transformed by oncogenic Ras and treated with Erlotinib. In this signature neuroregulin 1 (Nrg1) is down regulated while Id2 and Slc30a1 are up-regulated. Knockdown of the individual p38 isoforms does not reverse this trend (Figure 4C). Similar results were seen with Dusp1 and Timp3 also part of the EGFR ablation signature (data not shown).
Figure 4.
p38α is activated in EGFR ablated keratinocytes in the presence of oncogenic Ras. EGFR wildtype and EGFR null primary keratinocytes were transduced with v-rasHa retrovirus and subsequently transfected with a non-silencing siRNA (NS) or an siRNA targeting the three p38 isoforms, p38α (α), p38β (β), and p38δ (δ). After 24hrs, EGFR wildtype cells were treated with an EGFR pharmacologic inhibitor for 14 hours. A, Immunoblots for phospho-p38 (p-p38), p38α, and p38δ. B, Proliferation (3H-thymidine incorporation) of cultured v-rasHa-transduced EGFR null or wildtype keratinocytes after 4hr pre-treatment with p38α selective inhibitor (SB203580) and 14hr treatment with Erlotinib. P-values for EGFR WT group and EGFR null group were 0.0434 and 0.0227 respectively. . C, Nrg1, Id2, and Slc30a1 mRNA expression by real time PCR in EGFR wildtype and EGFR null keratinocytes. Columns, mean value of triplicate determinations, bars, SE.
p38 activation in conjunction with oncogenic Ras transformation is detected in human keratinocytes
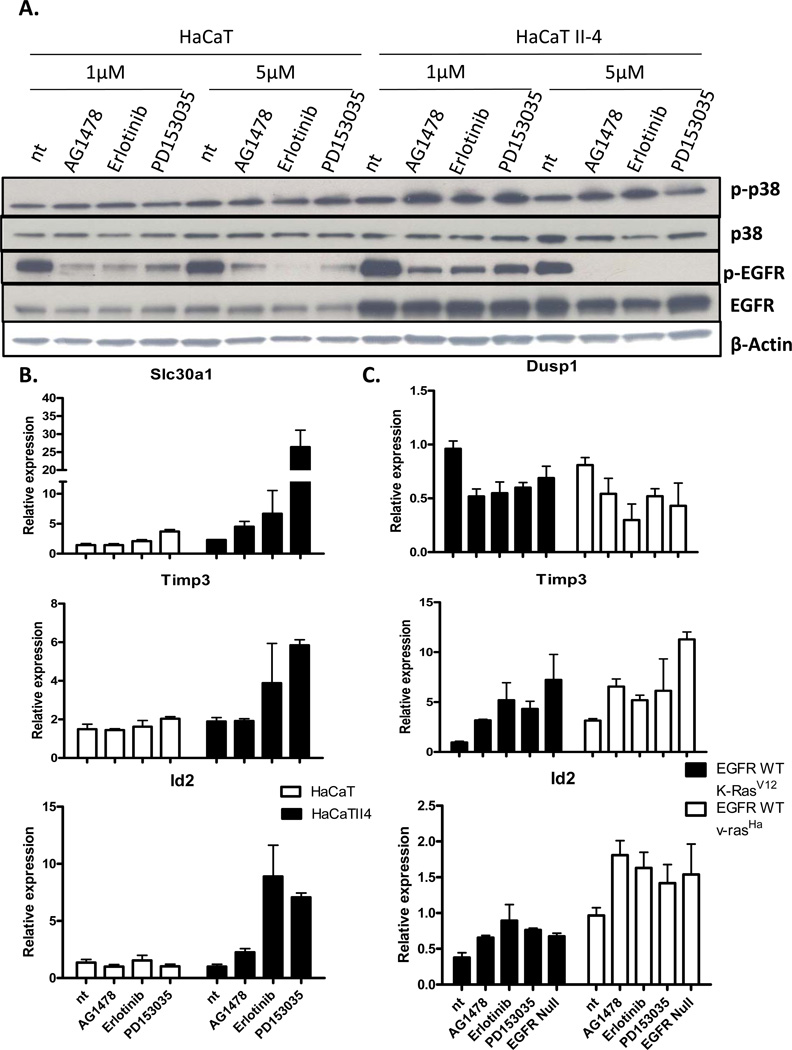
To confirm that changes in the v-rasHa-EGFR signature detected in mouse keratinocytes were relevant to human keratinocytes, HaCaT and oncogenic RAS transformed HaCaT II-4 cells (26) were employed. As seen in mouse keratinocytes, phospho-p38 is increased in the HaCaT II-4 cells when EGFR is pharmacologically abrogated (Figure 5A), but this response is not seen in the parental HaCaT cells. This result is consistent with previous finding in primary human keratinocytes where pharmacological inhibition of EGFR did not affect the level of phospho-p38 (27). Furthermore, signature v-rasHa-EGFR gene expression changes such as upregulation of Slc30a1, Id2 and Timp3 are increased upon EGFR inhibitor treatment specifically in the HaCaT II-4 cells (Figure 5B). K-RAS mutations are more commonly found in NSCLC than H-RAS mutations. To ensure that our v-rasHa-EGFR ablation signature was not limited to v-rasHa activation, but applied to K-rasV12 activation as well, we infected mouse keratinocytes with retroviruses expressing either K-rasV12 or v-rasHa (Supplemental Figure 2) and treated them with EGFR inhibitors. Gene expression changes seen in the v-rasHa-EGFR signature were also observed in the K-rasV12 expressing cells when inhibited for EGFR. Such gene expression changes seen were up-regulation of Timp3 and Id2 and down-regulation of Dusp1 (Figure 5C).
Figure 5.
Ras and EGFR abrogation dependent activation of p38 occurs in human keratinocytes. A, Immunoblots were performed for total and phospho-p38 (p-p38), and total and phospho-EGFR (p-EGFR) in HaCaT cells and HaCaT II-4 cells which have an activated H-Ras. B, Slc30a1, Id2, and Timp3 expression (real time PCR) in HaCaT and HaCaT II-4 cells treated with EGFR inhibitors. C, Expression of Dusp1, Timp3 and Id2 (real time PCR) in EGFR wildtype and EGFR null cells transduced with either K-rasV12 or v-rasHa and treated with inhibitors for EGFR. Columns, mean of triplicate determination, bars, SE.
Discussion
It is now well established that activating mutations in one of the RAS alleles is a negative prognostic factor for response to EGFR ablation therapy in multiple epithelial cancer types (9, 28, 29). We used an established mouse model for oncogenic v-rasHa- induced squamous cancer to profile gene expression changes that might illuminate genes or pathways that define Ras signaling in the absence of EGFR and therefore be potentially relevant to therapy. To narrow our analysis we required that both acute pharmacological ablation and chronic genetic ablation of EGFR be included and the genes limited to those recognized under the comprehensive map of the EGFR pathway (25). With these restrictions we were able to identify a 25 gene signature for further pathway analysis. By performing the same analysis on cells treated identically but without v-rasHa transduction, a distinct 19 gene signature was generated. The absence of overlap in the two gene lists reveals the importance of Ras signaling in cells lacking EGFR signaling. The study also revealed differences in gene expression and ontologies among chronic genetic EGFR ablation that may model extended human drug exposure and acute pharmacological ablation consistent with initiation of treatment. Several unexpected results became evident from this experimental design. In the absence of EGFR signaling, Ras activation has substantial influence on the expression of genes in ontologies for ion channels and membrane transporters. Further, pathway analysis and biochemical studies indicate that activation of p38α is a response to both acute and chronic abrogation of EGFR signaling under conditions of Ras activation. This was also observed in oncogenic RAS transduced human keratinocytes and in tumor grafts of v-rasHa transformed mouse keratinocytes. Changes in membrane ion transporters and p38α signaling could influence the therapeutic response to EGFR ablation therapy in tumors driven by oncogenic RAS.
Because these studies were performed on normal and v-rasHa- transformed keratinocytes, they may be most relevant to NSCLC and HNSCC patients with similar lining epithelia, but the biology is analogous in CRC. RAS mutations range from 5–40% in these tumor sites depending on ethnicity and smoking history among other factors (28–30). Whereas H-RAS mutations are more common in HNSCC, K-RAS mutations are more common in NSCLC. A sampling of our signature changes indicates similar results are obtained with either mutant RAS allele. These inherent differences of cancers with and without RAS mutations and the correlation to responsiveness with EGFR targeted therapy is mirrored in the distinct gene expression profiles documented in our model system using mouse keratinocytes ablated for EGFR signaling in the presence or absence of mutant ras. We noted distinct ontologies when oncogenic Ras was introduced into cells with chronically ablated EGFR (null keratinocytes) or acutely inhibited EGFR signaling (Erlotinib). Genes involved in cell cycle and transcriptional regulation are reduced with chronic depletion of EGFR while acute inhibition is characterized by up-regulation of transporters and stress related genes. Among the latter are increased expression of Ryanodine receptor and Atp2b3 (Pmca3) that are critical for Ca2+ transport and Ca2+ intracellular homeostasis. Over-expression of Atp2b3 increases the export of intracellular calcium; (31), thus lowering intracellular Ca2+ levels, a change that has been associated with chemoresistance (32, 33). It is well documented that altered calcium signaling in cancer cells contributes to tumor proliferation and progression, and molecules in the family of Atp2b3 participate in these alterations (34). Indeed, previous gene expression profiling of human lung cancer cell lines, validated on resected adenocarcinomas and related to clinical response, revealed a 180 gene signature predictive of response to Erlotinib that included multiple calcium signaling proteins including CaM kinase and Atp2c1, a member of the Atp2b3 family (35). These same genes are represented in a 26 gene signature predicting response to cetuximab in human colon cancer (36). However, from the available data the concordance with our findings cannot be confirmed.
Pathway analysis of gene expression changes revealed that inhibition of EGFR in cells with wildtype Ras proteins intercepted pathways centering on nodes for Erk and IL-1β signaling. These pathways would be relevant for normal tissues exposed to EGFR inhibitors for cancer therapy and could be associated with the acneiform skin rash so prevalent in these patients. Activation of the MAPK pathway is a feature of the skin rash in patients receiving Erlotinib for metastatic breast cancer (37). Similarly, anti-IL-1 therapy reduces the skin eruption in a mouse model for the human rash (38). We identified a unique activation of p38α as a consistent response in Ras transformed mouse and human keratinocytes after either acute or chronic EGFR ablation. This is associated with a consistent decrease in expression of Dusp1, the primary phosphatase responsible for deactivating p38α. Previous studies have indicated distinct functions for the four members of the p38 family. p38α contributes to multiple cellular responses and can mediate inflammation, proliferation, and cellular differentiation depending on cell type and context (39, 40). Activation of p38α is also associated with cancer cell proliferation (41–43) and invasion (44). p38α antagonists are under study as therapy for both inflammatory and neoplastic diseases alone or in combination with other targets (40, 42, 45, 46). In human keratinocytes p38α activation is required for migration on collagen 1 and release of MMP13 in RAS transformed keratinocytes (47). Thus, activation of p38α might worsen the response to EGFR ablation therapy in Ras driven tumors, and this has been observed in some cases. Our in vitro studies confirm that inhibiting p38 in oncogenic Ras/EGFR ablated keratinocytes reduces cell proliferation substantially suggesting a combination of EGFR and p38α inhibition could be clinically useful under similar circumstances.
In this study we have been able to define gene expression changes in keratinocytes before and after anti-EGFR therapy in a setting of potential therapeutic success or failure dependent on the presence of a ras oncogene. As far as we can tell only two previous studies have addressed tumor gene expression before and after treatment, and these were conducted with human metastatic breast cancers or breast cancer cell lines (48, 49). The large array platform we employed and the stringent requirements we used to restrict the analysis to genes only associated with EGFR signaling and consistent among multiple EGFR antagonists were designed to illuminate the most relevant gene changes attributable to therapy in a squamous tissue. We also incorporated both acute and chronic EGFR inhibition in our requirements. Despite the restrictions we imposed to get the most specific and relevant set of markers possible, expression changes in Serpine1, Tenascin C, Cdc6, Atp2b3, Foxd3 in our signatures were concordant with the same or closely related genes identified as the top changes in the breast cancer analysis. Of particular interest was the common finding of three minichromosome maintenance protein genes (MCM3, 6, and 7) commonly downregulated in our EGFR null gene list (Supplemental Table 3D) and breast cancer cells sensitive to lapatinib (53). These proteins are critical for DNA replication and cell cycle progression, ontologies commonly found suppressed by chronic EGFR ablation. Our approach offered an opportunity for the first time to explore functional pathways associated with the potential for treatment success or failure. We have identified alterations in calcium balance and p38 signaling as changes unique to oncogenic Ras transformation in the setting of EGFR inhibition and thus possible targets to overcome treatment resistance. With therapeutics now available to target each of these alterations, future studies can address the potential efficacy of combinations to achieve full tumor inhibition.
Statement of Translational Relevance.
A major advance in targeted cancer therapy was accomplished by the introduction of EGFR antagonists for the treatment of lung, head and neck and colon cancer. However, efficacy is limited due to the frequent presence of RAS gene mutations in these tumor types, an adverse skin eruption, and the development of resistance. This report utilizes a well described model of oncogenic Ras transformed keratinocytes, gene expression profiling and a highly restrictive analytic approach to identify unique expression signatures that define the influence of activated Ras on acute and chronic EGFR ablation therapy in a squamous tumor type. Combined with biochemical studies, the analysis has revealed several previously unrecognized functional alterations unique to the Ras-EGFR ablation interaction that could be targets for combination therapy to overcome therapeutic failure.
Supplementary Material
Acknowledgement
The authors wish to thank Joseph Riss, Aleksandra Michalowski and Richard Simon for advice on establishing the protocol and analyzing the data, and Marta Custer for care of the mouse colony.
This work was supported by the intramural program of the Center for Cancer Research, National Cancer Institute.
Footnotes
The authors have no conflict of interest to declare.
References
- 1.Lurje G, Lenz HJ. EGFR signaling and drug discovery. Oncology. 2009;77:400–410. doi: 10.1159/000279388. [DOI] [PubMed] [Google Scholar]
- 2.Iyer R, Bharthuar A. A review of erlotinib--an oral, selective epidermal growth factor receptor tyrosine kinase inhibitor. Expert Opin Pharmacother. 2010;11:311–320. doi: 10.1517/14656560903551283. [DOI] [PubMed] [Google Scholar]
- 3.Hartmann JT, Haap M, Kopp HG, Lipp HP. Tyrosine kinase inhibitors - a review on pharmacology, metabolism and side effects. Curr Drug Metab. 2009;10:470–481. doi: 10.2174/138920009788897975. [DOI] [PubMed] [Google Scholar]
- 4.Stewart DJ. Tumor and host factors that may limit efficacy of chemotherapy in non-small cell and small cell lung cancer. Crit Rev Oncol Hematol. 2010;75:173–234. doi: 10.1016/j.critrevonc.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chau I, Cunningham D. Treatment in advanced colorectal cancer: what, when and how? Br J Cancer. 2009;100:1704–1719. doi: 10.1038/sj.bjc.6605061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 7.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 8.Linardou H, Dahabreh IJ, Kanaloupiti D, Siannis F, Bafaloukos D, Kosmidis P, et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008;9:962–972. doi: 10.1016/S1470-2045(08)70206-7. [DOI] [PubMed] [Google Scholar]
- 9.Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens SL, Herbst RS, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 10.Zhu CQ, da Cunha Santos G, Ding K, Sakurada A, Cutz JC, Liu N, et al. Role of KRAS and EGFR as biomarkers of response to erlotinib in National Cancer Institute of Canada Clinical Trials Group Study. BR.21. J Clin Oncol. 2008;26:4268–4275. doi: 10.1200/JCO.2007.14.8924. [DOI] [PubMed] [Google Scholar]
- 11.Agero AL, Dusza SW, Benvenuto-Andrade C, Busam KJ, Myskowski P, Halpern AC. Dermatologic side effects associated with the epidermal growth factor receptor inhibitors. J Am Acad Dermatol. 2006;55:657–670. doi: 10.1016/j.jaad.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 12.Lacouture ME. Mechanisms of cutaneous toxicities to EGFR inhibitors. Nat Rev Cancer. 2006;6:803–812. doi: 10.1038/nrc1970. [DOI] [PubMed] [Google Scholar]
- 13.Li T, Perez-Soler R. Skin toxicities associated with epidermal growth factor receptor inhibitors. Target Oncol. 2009;4:107–119. doi: 10.1007/s11523-009-0114-0. [DOI] [PubMed] [Google Scholar]
- 14.Threadgill DW, Dlugosz AA, Hansen LA, Tennenbaum T, Lichti U, Yee D, et al. Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science. 1995;269:230–234. doi: 10.1126/science.7618084. [DOI] [PubMed] [Google Scholar]
- 15.Hansen LA, Woodson RL, 2nd, Holbus S, Strain K, Lo YC, Yuspa SH. The epidermal growth factor receptor is required to maintain the proliferative population in the basal compartment of epidermal tumors. Cancer Res. 2000;60:3328–3332. [PubMed] [Google Scholar]
- 16.Lichti U, Anders J, Yuspa SH. Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat Protoc. 2008;3:799–810. doi: 10.1038/nprot.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cataisson C, Ohman R, Patel G, Pearson A, Tsien M, Jay S, et al. Inducible cutaneous inflammation reveals a protumorigenic role for keratinocyte CXCR2 in skin carcinogenesis. Cancer Res. 2009;69:319–328. doi: 10.1158/0008-5472.CAN-08-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hlaing M, Spitz P, Padmanabhan K, Cabezas B, Barker CS, Bernstein HS. E2F-1 regulates the expression of a subset of target genes during skeletal myoblast hypertrophy. J Biol Chem. 2004;279:43625–43633. doi: 10.1074/jbc.M408391200. [DOI] [PubMed] [Google Scholar]
- 20.Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- 21.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cataisson C, Pearson AJ, Torgerson S, Nedospasov SA, Yuspa SH. Protein kinase C alpha-mediated chemotaxis of neutrophils requires NF-kappa B activity but is independent of TNF alpha signaling in mouse skin in vivo. J Immunol. 2005;174:1686–1692. doi: 10.4049/jimmunol.174.3.1686. [DOI] [PubMed] [Google Scholar]
- 25.Oda K, Matsuoka Y, Funahashi A, Kitano H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol. 2005;1:2005. doi: 10.1038/msb4100014. 0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mueller MM, Peter W, Mappes M, Huelsen A, Steinbauer H, Boukamp P, et al. Tumor progression of skin carcinoma cells in vivo promoted by clonal selection, mutagenesis, and autocrine growth regulation by granulocyte colony-stimulating factor and granulocyte-macrophage colony-stimulating factor. Am J Pathol. 2001;159:1567–1579. doi: 10.1016/S0002-9440(10)62541-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pastore S, Mascia F, Mariotti F, Dattilo C, Mariani V, Girolomoni G. ERK1/2 regulates epidermal chemokine expression and skin inflammation. J Immunol. 2005;174:5047–5056. doi: 10.4049/jimmunol.174.8.5047. [DOI] [PubMed] [Google Scholar]
- 28.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 29.Barton S, Starling N, Swanton C. Predictive molecular markers of response to epidermal growth factor receptor(EGFR) family-targeted therapies. Curr Cancer Drug Targets. 2010;10:799–812. doi: 10.2174/156800910793357925. [DOI] [PubMed] [Google Scholar]
- 30.Hardisson D. Molecular pathogenesis of head and neck squamous cell carcinoma. Eur Arch Otorhinolaryngol. 2003;260:502–508. doi: 10.1007/s00405-003-0581-3. [DOI] [PubMed] [Google Scholar]
- 31.Domi T, Di Leva F, Fedrizzi L, Rimessi A, Brini M. Functional specificity of PMCA isoforms? Ann N Y Acad Sci. 2007;1099:237–246. doi: 10.1196/annals.1387.043. [DOI] [PubMed] [Google Scholar]
- 32.Schrodl K, Oelmez H, Edelmann M, Huber RM, Bergner A. Altered Ca2+-homeostasis of cisplatin-treated and low level resistant non-small-cell and small-cell lung cancer cells. Cell Oncol. 2009;31:301–315. doi: 10.3233/CLO-2009-0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Padar S, van Breemen C, Thomas DW, Uchizono JA, Livesey JC, Rahimian R. Differential regulation of calcium homeostasis in adenocarcinoma cell line A549 and its Taxol-resistant subclone. Br J Pharmacol. 2004;142:305–316. doi: 10.1038/sj.bjp.0705755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkash J, Asotra K. Calcium wave signaling in cancer cells. Life Sci. 2010;87:587–595. doi: 10.1016/j.lfs.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balko JM, Potti A, Saunders C, Stromberg A, Haura EB, Black EP. Gene expression patterns that predict sensitivity to epidermal growth factor receptor tyrosine kinase inhibitors in lung cancer cell lines and human lung tumors. BMC Genomics. 2006;7:289. doi: 10.1186/1471-2164-7-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balko JM, Black EP. A gene expression predictor of response to EGFR-targeted therapy stratifies progression-free survival to cetuximab in KRAS wild-type metastatic colorectal cancer. BMC Cancer. 2009;9:145. doi: 10.1186/1471-2407-9-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan EH, Ramlau R, Pluzanska A, Kuo HP, Reck M, Milanowski J, et al. A multicentre phase II gene expression profiling study of putative relationships between tumour biomarkers and clinical response with erlotinib in non-small-cell lung cancer. Ann Oncol. 2010;21:217–222. doi: 10.1093/annonc/mdp520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Surguladze D, Deevi D, Claros N, Corcoran E, Wang S, Plym MJ, et al. Tumor necrosis factor-alpha and interleukin-1 antagonists alleviate inflammatory skin changes associated with epidermal growth factor receptor antibody therapy in mice. Cancer Res. 2009;69:5643–5647. doi: 10.1158/0008-5472.CAN-09-0487. [DOI] [PubMed] [Google Scholar]
- 39.Ashwell JD. The many paths to p38 mitogen-activated protein kinase activation in the immune system. Nat Rev Immunol. 2006;6:532–540. doi: 10.1038/nri1865. [DOI] [PubMed] [Google Scholar]
- 40.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 41.Comes F, Matrone A, Lastella P, Nico B, Susca FC, Bagnulo R, et al. A novel cell type-specific role of p38alpha in the control of autophagy and cell death in colorectal cancer cells. Cell Death Differ. 2007;14:693–702. doi: 10.1038/sj.cdd.4402076. [DOI] [PubMed] [Google Scholar]
- 42.van Houdt WJ, de Bruijn MT, Emmink BL, Raats D, Hoogwater FJ, Borel Rinkes IH, et al. Oncogenic K-ras activates p38 to maintain colorectal cancer cell proliferation during MEK inhibition. Cell Oncol. 2010;32:245–257. doi: 10.3233/CLO-2010-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Halawani D, Mondeh R, Stanton LA, Beier F. p38 MAP kinase signaling is necessary for rat chondrosarcoma cell proliferation. Oncogene. 2004;23:3726–3731. doi: 10.1038/sj.onc.1207422. [DOI] [PubMed] [Google Scholar]
- 44.Junttila MR, Ala-Aho R, Jokilehto T, Peltonen J, Kallajoki M, Grenman R, et al. p38alpha and p38delta mitogen-activated protein kinase isoforms regulate invasion and growth of head and neck squamous carcinoma cells. Oncogene. 2007;26:5267–5279. doi: 10.1038/sj.onc.1210332. [DOI] [PubMed] [Google Scholar]
- 45.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 46.Yong HY, Koh MS, Moon A. The p38 MAPK inhibitors for the treatment of inflammatory diseases and cancer. Expert Opin Investig Drugs. 2009;18:1893–1905. doi: 10.1517/13543780903321490. [DOI] [PubMed] [Google Scholar]
- 47.Li W, Nadelman C, Henry G, Fan J, Muellenhoff M, Medina E, et al. The p38-MAPK/SAPK pathway is required for human keratinocyte migration on dermal collagen. J Invest Dermatol. 2001;117:1601–1611. doi: 10.1046/j.0022-202x.2001.01608.x. [DOI] [PubMed] [Google Scholar]
- 48.Yang SX, Simon RM, Tan AR, Nguyen D, Swain SM. Gene expression patterns and profile changes pre- and post-erlotinib treatment in patients with metastatic breast cancer. Clin Cancer Res. 2005;11:6226–6232. doi: 10.1158/1078-0432.CCR-05-0270. [DOI] [PubMed] [Google Scholar]
- 49.Hegde PS, Rusnak D, Bertiaux M, Alligood K, Strum J, Gagnon R, et al. Delineation of molecular mechanisms of sensitivity to lapatinib in breast cancer cell lines using global gene expression profiles. Mol Cancer Ther. 2007;6:1629–1640. doi: 10.1158/1535-7163.MCT-05-0399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.