Abstract
Many higher plants are polysomatic whereby different cells possess variable amounts of nuclear DNA. The conditional triggering of endocycles results in higher nuclear DNA content (C value) that in some cases has been correlated to increased cell size. While numerous multicolored fluorescent protein (FP) probes have revealed the general behavior of the nucleus and intranuclear components, direct visualization and estimation of changes in nuclear-DNA content in live cells during their development has not been possible. Recently, monomeric Eos fluorescent protein (mEosFP) has emerged as a useful photoconvertible protein whose color changes irreversibly from a green to a red fluorescent form upon exposure to violet-blue light. The stability and irreversibility of red fluorescent mEosFP suggests that detection of green color recovery would be possible as fresh mEosFP is produced after photoconversion. Thus a ratiometric evaluation of the red and green forms of mEosFP following photoconversion could be used to estimate production of a core histone such as H2B during its concomitant synthesis with DNA in the synthesis phase of the cell cycle. Here we present proof of concept observations on transgenic tobacco (Nicotiana tabacum) Bright Yellow 2 cells and Arabidopsis (Arabidopsis thaliana) plants stably expressing H2B::mEosFP. In Arabidopsis seedlings an increase in green fluorescence is observed specifically in cells known to undergo endoreduplication. The detection of changes in nuclear DNA content by correlating color recovery of H2B::mEosFP after photoconversion is a novel approach involving a single FP. The method has potential for facilitating detailed investigations on conditions that favor increased cell size and the development of polysomaty in plants.
Polysomaty, defined as the variability in nuclear-DNA content that occurs among different cells existing in a single body, occurs commonly in higher plants (Berger, 1941 and refs. therein). A major mechanism, but not the only one, involved in the development of polysomaty is endoreduplication, wherein repeated cycles of DNA synthesis take place without the intervention of a cell division phase (Kondorosi et al., 2000; Walker et al., 2000; Sugimoto-Shirasu and Roberts, 2003; del Mar Castellano et al., 2004; Bramsiepe et al., 2010). Consequently, endoreduplicated cells possess nuclear-DNA content (C value) that might be several folds that of the diploid genome for the specific species. Higher C values have been suggested to increase mRNA and protein production in a cell and generally correlate with large rapidly growing cells (Donnelly et al., 1999; Kondorosi et al., 2000; Sugimoto-Shirasu and Roberts, 2003; Jakoby and Schnittger, 2004; Jovtchev et al., 2006; Lee et al., 2009). These correlations have been particularly apparent in the rhl1/hyp7 mutants of Arabidopsis (Arabidopsis thaliana) that affect the average cell size (Sugimoto-Shirasu et al., 2005) and the try, kak2, rfi, and gl3 mutants where leaf epidermal trichomes have altered size and branching (Hülskamp et al., 1994). Flow cytometry (Zhang et al., 2005; Castro et al., 2007), and morphometric estimates of nuclei stained with the fluorescent AT nucleic-acid-specific dye, 4′6-diamidino-2-phenylindole provide useful parameters for estimating C values (Inzé and De Veylder, 2006; Breuer et al., 2010). Notably these methods are informative only after endoreduplication has occurred and produced an increase in nuclear-DNA content of a cell. The methods do not provide any information on the process as it happens and therefore are not very useful for pinpointing cells that have shifted into an endoreduplication cycle during their development. Consequently, despite the high incidence of endoreduplication in plants and commendable progress in identifying molecular players involved in the regulation of endocycles (John and Qi, 2008; Breuer et al., 2010; Ishida et al., 2010) we are still far from defining physiological conditions that trigger a switch from a mitotic cycle to an endocycle.
In contrast to our inability to follow the phenomenon of endoreduplication in real time, considerable advances have been made in imaging nuclei and intranuclear components through the use of multicolored fluorescent proteins (FPs; Table I). While highlighting their target clearly the cyan, green, yellow, and red fluorescent probes used in these studies (Table I) are limited in their applications since their colors cannot be switched on or altered as and when required.
Table I. Some useful multicolored FP probes for visualizing the nucleus and subnuclear components in living plant cells.
C, Cyan; G, green; Y, yellow; R, red; X, many different colors.
| Target | FP Color | Fusion with | Reference |
| Nucleus | G | NLS-sGFP::GUS | Grebenok et al. (1997) |
| Minimal NLS from c2 polypeptide of tobacco, fusion to GUS prevents size-related diffusion from the nucleus | Chytilova et al. (1999) | ||
| Y | Histone H2B | ||
| C-terminal fusion to the full-length cDNA of the Arabidopsis H2B gene | Boisnard-Lorig et al. (2001) | ||
| G/R | PopP2 | ||
| N-terminal fusion to the full-length genomic PopP2 gene of Ralstonia solanacearum | Deslandes et al. (2003) | ||
| G/Y | HTA6 (histone H2A) | ||
| C-terminal fusion to the full-length cDNA of the Arabidopsis HTA6 gene | Zhang et al. (2005) | ||
| Y/C | pAtHB8-NLS-YFP and pAtHB8-CFP-NLS promoter region of AtHB8 (1,997 bp upstream of ORF) and SV40 NLS | Sawchuk et al. (2007) | |
| G | LeMAF1-GFP-LeMAF1 | ||
| Full-length cDNA of the Lycopersicon esculentum (cv VFNT cherry) MFP1 associated factor1 (MAF1) gene | Gindullis et al. (1999) | ||
| G | SM40::GFP | ||
| C-terminal fusion of Mammalian simian virus 40 large T-antigen nuclearlocalization signal (amino acid residues 126–132) | Chiu et al. (1996) | ||
| Nuclear envelope | G | AtRAN-GAP1 | |
| C-terminal fusion to the full-length cDNA of Arabidopsis RanGTPase-activating protein1 (RAN-GAP1) | Rose and Meier (2001) | ||
| G | RAN-GAP1 and RAN-GAP2 | ||
| C-terminal fusion to the full-length cDNA of Arabidopsis RanGTPase-activating protein1(RAN-GAP1) and RAN-GAP2 | Pay et al. (2002) | ||
| G | Lamin B receptor | ||
| C-terminal fusion to the first 238 amino acids of the Homo sapiens Lamin B receptor | Irons et al. (2003) | ||
| G | AtSad1a and AtSad1b | ||
| C-terminal fusion to the full-length cDNA of Arabidopsis Sad1a and b genes | Van Damme et al. (2004) | ||
| G | MOS3 | ||
| C-terminal fusion to the full-length cDNA of Arabidopsis MOS3 (MOFIDIER OF SNC1) | Zhang and Li (2005) | ||
| G | AtSUN1 (Sad1/UNC84) | Graumann et al. (2010) | |
| AtSUN2 | Graumann and Evans (2011) | ||
| Nuclear pores | G | Nup136/Nup1 | |
| C-terminal fusion to the genomic and full-length cDNA of Arabidopsis Nup136 | Tamura et al. (2010); Tamura and Hara-Nishimura (2011) | ||
| Nucleolus | G | AtFbr1::smGFP | |
| C-terminal fusion to the full-length cDNA of Arabidopsis FIBRILLARIN1 | Pih et al. (2000) | ||
| G | Nop10 (At2g20490) | Lorković et al. (2004) | |
| Cajal bodies | G | U2B” (At2g30260) | Boudonck et al. (1999) |
| G | ORF3 | Kim et al. (2007) | |
| AtCoilin | Kim et al. (2007) | ||
| Nuclear bodies/speckles | G/R | Arabidopsis CRYPTOCHROME 2 (CRY2-At1g04400) | Yu et al. (2009) |
| X | Different SR proteins | Lorković et al. (2004) | |
| G | COP1 (At2g32950) | Wang et al. (2001); Yang et al. (2001) | |
| Chromatin | G | LIKE HETEROCHROMATIN PROTEIN1 (LHP1: At5g17690) | Libault et al. (2005); Zemach et al. (2006) |
| Heterochromatin | G | Heterochromatin protein 1 (HP1γ human) | Fass et al. (2002) |
| Euchromatin | G | Arabidopsis histone deacetylase 1 (AF014824) | Fong et al. (2006) |
| Centromere | G | Polydactyl zinc finger (PZF) DNA binding domains | Lindhout et al. (2007) |
| G | Histone3 (HTR12; At1g01370) | Fang and Spector (2005) | |
| C | Histone 2B (HTB1:At1g07790) | Fang and Spector (2005) | |
| Y | Histone H3 (CENH3) | Moraes et al. (2011); Ingouff et al. (2007, 2010) |
Recently a number of optical highlighter proteins that can be photoinduced, photoconverted, and photoswitched between two fluorescent states have become available and introduce a very high degree of precision into live-cell imaging (Shaner et al., 2007). A monomeric green to red photoconvertible form of Eos fluorescent protein (EosFP) (Wiedenmann et al., 2004) is especially useful for application in plants as it provides the ability to differentially color and track a single organelle within a population, as well as follow endomembrane and cytoskeletal dynamics over time (Mathur et al., 2010). An important property of monomeric EosFP (mEosFP) is the high stability and irreversibility of its red fluorescent photoconverted form. Thus, if green fluorescence of a target organelle or cell increases and reappears after photoconversion it can be attributed largely to fresh green fluorescent mEosFP. Therefore, in conjunction with an appropriate nuclear protein or nuclear localization signal sequence (NLS), we hypothesized that this singular property of mEosFP could be used to detect a change in green fluorescence of a red-colored nucleus following DNA synthesis. Here, we have used histone H2B as our target nuclear protein.
Histones constitute some of the most conserved nuclear proteins within the eukaryotic cell. As the basic core for DNA packaging in a nucleosome, histone turnover and DNA replication are spatiotemporally linked during the synthesis (S) phase of the eukaryotic cell cycle (Robbins and Borun, 1966; Hardin et al., 1967; Hnilica, 1972). Moreover, different histones have already been fused to FPs for observing intranuclear dynamics in diverse organisms (Boisnard-Lorig et al., 2001; Kimura and Cook, 2001; Kimura, 2005; Foudi et al., 2009). In plants, the histones H2A/HTA6 (Zhang et al., 2005), H2B (Boisnard-Lorig et al., 2001; Adachi et al., 2011), and H3 (Ingouff et al., 2007, 2010) have been used successfully (Table I). Arabidopsis plants stably expressing H2B::(x)FP fusions have been created and display normal cell functions and plant development (Boisnard-Lorig et al., 2001; Van Bruaene et al., 2003; Adachi et al., 2011).
Here we report the development and characterization of green to red photoconvertible H2B::mEosFP for plants and use it to demonstrate that green color recovery after photoconversion can be used for detecting an increase in H2B production and correlated with increased nuclear DNA content. We observe that while the color of certain nuclei remains red, other nuclei exhibit a gradual shift toward a green fluorescent state. Whereas red fluorescent nuclei are limited to cells that do not undergo cell division or endoreduplication the shift toward green fluorescence is always found in the nuclei of cells that are known to undergo endoreduplication. The use of color recovery after photoconversion in conjunction with the novel, green to red photoconvertible H2B::mEosFP probe now present in stable transgenic Arabidopsis and Bright Yellow 2 (BY2) cell lines constitutes a novel approach for the visualization and correlative estimation of changes in nuclear DNA content in living cells.
RESULTS AND DISCUSSION
Photoconvertible H2B::mEosFP Fusion Allows Differential Labeling of Nuclei and Chromosomes in Living Plant Cells
Different FPs fused to H2B have been used for understanding nuclear behavior in different eukaryotes. The creation of a C-terminal fusion of Arabidopsis H2B cDNA (At3g45980) with a mEosFP (Wiedenmann et al., 2004) resulted in the H2B::mEosFP fusion reported here. Transient overexpression of H2B::mEosFP driven by a cauliflower mosaic virus (CaMV) 35S promoter in tobacco (Nicotiana tabacum) leaves highlighted nuclei in a fluorescent green color. The green nuclear fluorescence readily changed to red following the exposure to violet-blue light for 10 s. Subsequently, several stable transgenic Arabidopsis and tobacco BY2 lines expressing H2B::mEosFP were generated. In the 30 independent Arabidopsis transgenic lines selected six lines exhibited slightly reduced growth in the T1 generation. Whereas subsequent generations of homozygous transgenic plants will be used for detailed comparisons with wild-type seedlings for uncovering T-DNA insertion-based and H2B overexpression-induced effects, the four independent insertion lines used in the experiments reported here exhibited normal germination and seedling development.
As one of the major reasons for creating photoconvertible H2B::mEosFP is to use it eventually for understanding the development of polysomaty, we first investigated the occurrence of this phenomenon in transgenic Arabidopsis plants. It is known that neighboring cells of the cotyledons and leaves have different shapes and sizes as well as different nuclear DNA content. This was apparent upon comparing the size of nuclei in guard cells with the nuclear size of pavement (Fig. 1A) and trichome cells (Fig. 1B). Whereas guard cells are recognized as terminally differentiated and known to maintain the conserved 2 C value, pavement cells can display C values of 4 C, 8 C, or 16 C and trichomes in Arabidopsis can have nuclear DNA content of up to 32 C (Galbraith et al., 1991; Melaragno et al., 1993; Sugimoto-Shirasu and Roberts, 2003; Ishida et al., 2008, 2010). High nuclear DNA content of 8 and 16 C is also encountered in large epidermal cells of floral sepals in Arabidopsis (Roeder et al., 2010; Adachi et al., 2011). A visual comparison of the size of the nuclei in guard cells versus the nuclei in enlarged cells of sepals (Fig. 1C) supported earlier correlations between nuclear size and endoploidy (Kondorosi et al., 2000; Sugimoto-Shirasu and Roberts, 2003). By contrast, cells surrounding the root apical meristem exhibited a narrower variation in nuclear sizes and could also be easily photoconverted (Fig. 1D). Since H2B is known to be recycled more in comparison to other histones in mammalian cells (Kimura and Cook, 2001) we assessed the possibility that photoconverted H2B::mEosFP might diffuse within large nuclei. Figure 1B shows localized photoconversion of a portion of a large trichome cell nucleus that was then observed over 4 h for the diffusion of the red fluorescent signal. The red region, as well as the surrounding circular, yellow fringe was maintained throughout this time and did not diffuse to create a yellow- or green-colored nucleus. This indicated that photoconverted molecules of H2B::mEosFP remained tightly bound and were not free to diffuse within the nucleus. Having established that polysomaty can be easily observed in Arabidopsis plants and that single nuclei can be labeled in living cells using this probe, we used tobacco BY2 cells to understand the stability of the red photoconverted form of this probe through the cell cycle.
Figure 1.
Representative nuclear size and colors in stably transformed cells expressing H2B::mEosFP. Nuclei are green fluorescent when mEosFP is in an unphotoconverted state, fluoresce red following complete photoconversion, or appear yellow fluorescent when partial photoconversion results in nearly equal numbers of green and red molecules of mEosFP. A, Different nuclear sizes indicating polysomaty are seen on a single leaf from transgenic Arabidopsis. Whereas nuclei in guard cells (arrowheads) present the diploid (2n) state with 2 C nuclear DNA content a neighboring pavement cell (asterisk) possesses a larger nucleus whose DNA content may be 4 C or 8 C due to endoreduplication. B, A leaf epidermal hair (trichome; asterisk) exhibits a larger nucleus in comparison to green fluorescent nonphotoconverted nuclei in other epidermal cells, suggesting higher nuclear DNA content value of 16 or 32 C in the trichome. Photoconversion of a small region of the trichome nucleus (red) and observing it over 4 h suggested that red H2B::mEosFP does not diffuse within the nucleus of nondividing, terminally differentiated cells. C, Floral sepal cells from a transgenic Arabidopsis plant exhibit a wide range of nuclear sizes; arrowheads point to 2 C nuclei in guard cells while larger epidermal cells exhibit significantly bigger nuclei (red, yellow; asterisks), apparently due to higher nuclear DNA content. D, Nuclei in the root apex display a regular size distribution in comparison to cells of the leaf epidermis. A short irradiation of 6 s with violet-blue light step is sufficient for substantial photoconversion but leaves residual green fluorescence (compare section 2 to 3). E, Cells from a stably transformed tobacco BY2 cell suspension line exhibit bright fluorescent nuclei. An entire nucleus (asterisk) or a small portion (arrowheads) can be photoconverted into a red fluorescent color. F, A single nucleus preparing to undergo mitosis shows clear chromosomes of which some have been photoconverted into red color. G, Differentially colored chromosomes in a cell (boxed in E) maintain their photoconverted state while passing through the M phase of the cell cycle (1, late metaphase; 2, early anaphase; 3, late anaphase). Size bars: A, B, C, E = 25 μm; D = 50 μm; F and G = 5 μm.
Differential Coloring of Nuclei and Chromosomes during the S-G2 Phase Is Maintained through One Mitotic Cycle
Tobacco BY2 cells undergo rapid division and constitute a well-accepted system for understanding cell-cycle-related events in plants (Nagata et al., 2003). In general, under optimal conditions, cells in an unsynchronized BY2 culture multiply 80- to 100-fold per week; doubling in about 13 h with an approximate 5-h S phase and a 2-h M phase (Kumagai-Sano et al., 2007). Stably transformed cells in different BY2 cell lines displayed large, bright-green fluorescent nuclei (Fig. 1E). An entire nucleus, or even a small portion of the nucleus could be photoconverted to display red fluorescence (Fig. 1E). Randomly selected nuclei were photoconverted and subjected to time-lapse imaging. Depending upon the time after subculture a varying number of cells progressed from the S-G2 phase (e.g. Fig. 1F) into the M phase. Figure 1E shows a single cell (boxed) where some of the chromosomes aligned at the metaphase plate. Within the next 60 min, the cell moved from metaphase into anaphase while maintaining differential coloring of the chromosomes (Fig. 1F).
Nuclear fluorescence in cells that divided and went into the next gap phase become diffuse and could not be followed into the subsequent cycle. Presently, we attribute this inability to the technical limitation of maintaining BY2 cells in a continually dividing state after they have been brought into a relatively stagnant situation on a glass slide. BY2 cells require constant shaking and contact with well-aerated cell-suspension medium for maintaining their division status. Under suboptimal conditions the G1 phase can be prolonged considerably. This appeared to happen in the case of BY2 cells being observed by us. Nevertheless, the dilution of the photoconverted form of the fusion protein due to the addition of fresh green H2B::mEosFP suggested that estimating the increase of green color in a red fluorescent nucleus could be used for detecting the onset of a new S phase. However, a change in the green to red ratio could also result from the degradation of red fluorescent mEosFP. Therefore we assessed the stability of the red form of H2B::mEosFP in living cells through observations on photoconverted nuclei in terminally differentiated guard cells possessing a 2C nuclear DNA content. The red fluorescence of these nuclei (n = 13) was very stable with an average brightness value ranging between 55 ± 9 on a 0 to 255 RGB color scale and a loss of less than 5% relative fluorescence over 24 h of observation (Supplemental Fig. S1). Establishing the stability of red fluorescent H2B::mEosFP in diploid guard cell nuclei suggested a time frame of 24 h at least for carrying out subsequent experiments aimed at observing changes in nuclear color in other cell types.
Red Fluorescent Nuclei Shift to a Green Fluorescent State in Tip-Growing Hair Cells
It has been established through flow cytometry and morphometric techniques that the rapid enlargement of many plant cells correlates with increased nuclear DNA content due to multiple rounds of endoreduplication. Here we photoconverted nuclei of tip-growing root and collet epidermal hairs that are known to undergo endoreduplication (Galbraith et al., 1991; Sugimoto-Shirasu et al., 2005; Ishida et al., 2008), for assessing the possibility of green fluorescence recovery following increased H2B::mEosFP production due to the onset of a fresh endocycle.
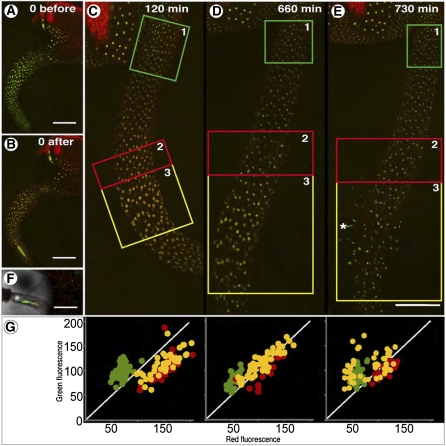
Seeds of transgenic Arabidopsis lines expressing H2B::mEosFP were germinated in specially designed thin chambers (“Materials and Methods” section). Photoconversion and long-term observations were initiated on emergent seedlings 3 d after imbibition (Fig. 2). At this stage the radical and a small portion of the hypocotyl had emerged out of the seed coat (Fig. 2A). Six cycles consisting of 30 s of irradiation with violet-blue light followed by a 15-s break were used to photoconvert the green fluorescent nuclei to red fluorescent nuclei (Fig. 2B). Subsequent time-lapse images taken every 30 min showed that by 120 min a major portion of the hypocotyl region had emerged. Since the nuclei in this region had not been photoconverted they appeared green (Fig. 2C, box 1). Red and green (RG) fluorescence measurements of nuclei (n = 117) in the seedling shown in Figure 2, on a 0 to 255 value scale, were divided into three populations (boxed regions 1, 2, and 3; Fig. 2, C–E). Nuclei in box 1 (Fig. 2C) belonged to the nonphotoconverted population and were predominantly green. Box 2 showed red photoconverted nuclei of the hypocotyl, whereas box 3 displayed photoconverted nuclei in the lower portion of the hypocotyl. This portion is also called the collet or collar region and represents the transition zone between the aerial shoot and the root. Epidermal cells of the collet region produce hair that undergo endoreduplication and elongate considerably through tip growth (E. Sliwinska, J. Mathur, and J.D. Bewley, unpublished data). Observations on the seedling at 730 min showed the emergence of several collet hairs (Fig. 2, E and F) whereas a comparative analysis of the three boxed regions (Fig. 2, C–E) showed a major shift in the color of the nuclei in region 3 (Fig. 2G). Internal controls such as the nonphotoconverted nuclei (green box) and autofluorescence of the seed coat (bright-red color in the left corner; Fig. 2, C and E) were used for ensuring that image acquisition parameters were maintained during the experiment (Supplemental Fig. S3).
Figure 2.
Estimation of green color recovery after photoconversion of nuclei in hypocotyl and collet region of a transgenic Arabidopsis seedling expressing H2B::mEosFP. A, Seedling before photoconversion at the beginning of the time series showing emerging root. Most of the hypocotyl region is still within the seed coat. B, Seedling immediately after six photoconversion cycles of 30 s each, whereupon all nuclei are colored red. C to E, Seedling after 120, 660, and 730 min of growth during which the elongating hypocotyl emerges completely out of the seed coat. Nuclei in the newly emerging tissue are not photoconverted and appear green. Three regions can be discerned: 1, green box showing fresh emergent hypocotyl cells with green nuclei; 2, red box with photoconverted nuclei in hypocotyl cells; 3, yellow box with photoconverted nuclei in the collet zone of cellular differentiation. F, Magnified view of a single collet hair cell (asterisk in E) exhibiting a large green nucleus. G, Scatter plot of RG fluorescence brightness values (on a scale of 0–255; 256 color code) for individual nuclei from boxes 1, 2, and 3 in C, D, and E. At 120 min the photoconverted and unconverted areas show a clear difference in nuclear color. At 660 min the color separation is still maintained although the color of some photoconverted nuclei exhibits a slight shift toward green. At 730 min a major green shift in nuclear color is observed and is limited to individual, slightly elongated nuclei in hairs extended from the collet region. Nonphotoconverted nuclei in the hypocotyl (green box) and the autofluorescent seed coat (left corner) were used as internal controls for ensuring that image acquisition parameters have been maintained during the experiment (Supplemental Fig. S3). Size bars: A to E = 200 μm; F = 50 μm.
We concluded that a shift toward green coloration suggesting the production of fresh H2B::mEosFP specifically in an endoreduplicating cell type could be correlated with the onset of an endocycle in a cell. Our investigations profited from the serendipitous observation that an asymmetric environment favors root hair elongation on one side of a seedling only. Observations on such seedlings allowed us to compare the behavior of nuclei in actively growing and relatively inactive cells of the same kind.
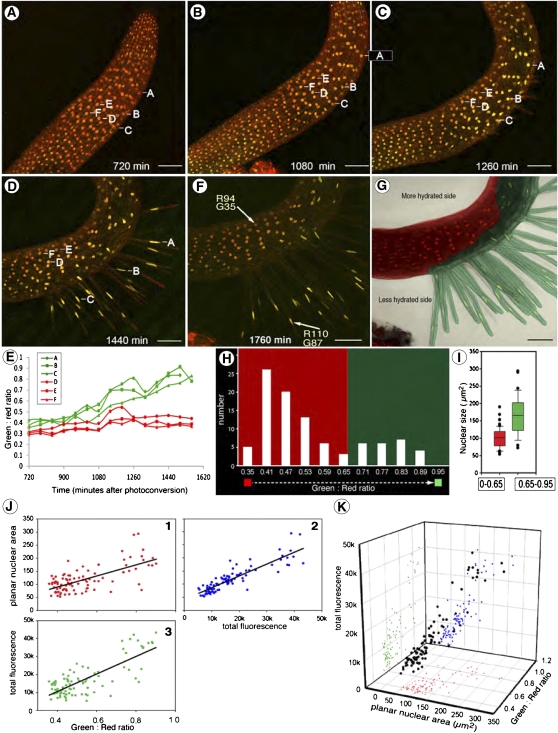
Asymmetric Hair Elongation in a Seedling Coincides with Increased Nuclear Size and Green-Shifted Fluorescence in Actively Growing Cells
Some of the seedlings in our live-imaging chambers developed hairs on only one side. A close inspection of the setup revealed that the moisture content on opposite sides of the seedling was different. While we did not measure the moisture content we followed nuclear behavior, increase in size, and the tip growth of collet and root hair cells (Fig. 3). In the representative time images of this seedling, six nuclei labeled A to F (Fig. 3A) were followed over time. As shown in Figure 3, A to D, nuclei A, B, and C belonged to cells that embarked on tip growth whereas cells containing D, E, and F did not develop into hairs. By 1,260 min (Fig. 3C) a clear difference in the color started becoming apparent whereas by 1,440 min nuclei exhibited both change in color and size (Fig. 3D). Plotting the green to red ratio for each nucleus over time (Fig. 3E) shows that around 720 min the color value for the six nuclei ranged between 0.3 and 0.45. Nuclei A, B, and C already showed a slightly higher green value in comparison to the others, suggesting that there might be increased H2B turnover already in these nuclei. The green values for all nuclei increased between 900 and 1,620 min but the largest increase occurred in nuclei A, B, and C. These three nuclei with increasing histone content, and by correlation increasing DNA content, belong to cells that exhibit active tip growth (Fig. 3D). In contrast cells that stayed relatively inactive, despite having the capability of carrying out tip growth, showed low green to red ratio. As shown in Figure 3, F to H the distribution of nuclei with increased green fluorescence divides the area of the seedling being visualized into two well-demarcated developmental regions (Fig. 3G). In concurrence with studies that have applied flow cytometry and nuclear morphometry to endoreduplicating root hair cells Figure 3I suggests an increased size for nuclei exhibiting higher green fluorescence.
Figure 3.
Confocal time-lapse imaging over 1,760 min of an Arabidopsis seedling expressing H2B::mEosFP and developing collet and root hairs asymmetrically due to differences in hydration levels on opposite sides of the seedling. A to D, Maximum intensity projections of z stacks taken at 720 (A), 1,080 (B), 1,260 (C), 1,440 (D), and 1,760 (E) min to assess color changes in nuclei. Six nuclei labeled A to F (section A) followed over time; nuclei A, B, and C belong to cells that exhibited rapid polar elongation whereas D, E, and F belong to cells that did not differentiate further. E, Color change in nuclei A to F (section A) calculated as the green to red ratio of the respective mean fluorescence values (0–255 value scale) plotted over time informs about the start time and progression through for an endocycle. F, Measurement of green (G) and red (R) fluorescence values in two visible nuclei (arrows) in the last image in the time series (1,760 min; F) suggests clear differences in color. G, Nuclei (n = 96) from epidermal and collet or root hair cells observed in F classed on the basis of their green to red ratio presented as a bar diagram. A value close to 0 represents a red and a ratio close to 1 a more green color. When considered overall the nuclei fall into two populations on the basis of their color. One population centers on green/red 0.47, and the other, separated by a gap of 0.65, around 0.77. H, A color overlay representation reflects the distribution of the two groups of different-colored nuclei with values below 0.65 depicted in red and above 0.65 overlaid in green. The seedling is divided into two asymmetrically developing zones. I, A box plot representing the two groups of different-colored nuclei with values below 0.65 depicted in red and above 0.65 depicted in green. The comparison of both populations shows that nuclei with ratios above 0.65 tend to be larger than nuclei below this ratio (P value < 0.00001). J, Establishing correlations between color change (green to red ratio) due to net increase in H2B::mEosFP content in nuclei, alteration in planer nuclear area, and the total nuclear fluorescence. Pairwise two-dimensional scatter plots showing planar nucleus area (in μm2; J1), the total nuclear fluorescence (J2), and the green to red ratio suggest a positive correlation between the three parameters (R2 J1 = 0.460; J2 = 0.711; J3 = 0.582). K, A combined three-dimensional scatter plot and multiregression analysis (ANOVA R2 = 0.8459) reaffirms a strong correlation of changes in the green to red ratio to other parameters.
Correlational analysis of the green to red ratio in photoconverted nuclei during development, change in size of nuclei, and alteration in the total brightness of individual nuclei was performed and is presented in Figure 3J, 1, 2, and 3 and further combined as a three-dimensional scatter plot (Fig. 3K). A multiple regression analysis using total fluorescence as the dependent variable and both green to red ratio and planar nuclear area as the independent variables returned an R-square value of 0.8459, indicating a significant correlation between the three parameters. Both independent variables contribute significantly to the prediction of total fluorescence, with P < 0.0001. The triparametric analysis clearly established that nuclei that showed a change in green to red ratio were also brighter in their total fluorescence (suggesting an increase in histone and DNA content) and had larger nuclear size. Whereas the differential sensitivity of detectors to green and red wavelengths can bias the value for total fluorescence the use of three independent variables and retracing them to the tissue zones used for obtaining the values (Supplemental Fig. S2) provides visual confirmation of the correlations.
The association between nuclear endoreduplication and the initiation and maintenance of tip growth in root hairs is well established and its conditional triggering as a developmental response of a living plant was confirmed through our experiments. Our approach involving color recovery after photoconversion was assessed further in the epidermal cells of the hypocotyl, another well-acknowledged endoreduplicating cell type.
Green Color Recovery after Photoconversion in Nuclei Coincides with Cell Elongation in Hypocotyl Cells
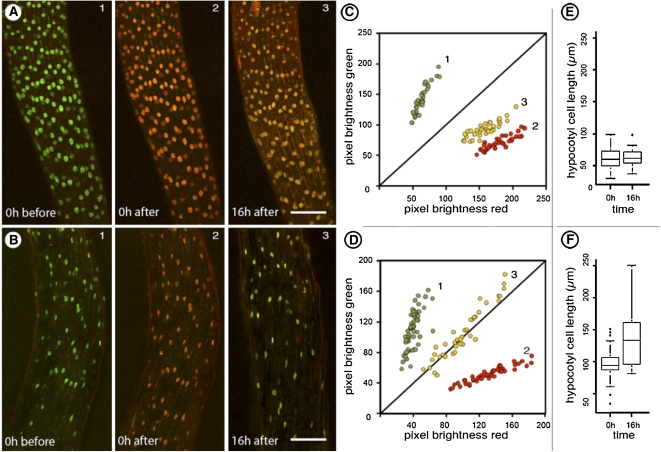
The hypocotyl of seedlings grown in dark becomes greatly elongated through a variable number of endoreduplication cycles (Melaragno et al., 1993; Barow, 2006). We used Arabidopsis seedlings to detect changes in color of the nuclei of hypocotyl cells while they elongated under dark conditions. Figure 4 presents representative observations made on seedlings grown in light before they were photoconverted and placed into light (Fig. 4A, 3) or into dark (Fig. 4B, 3). Observations taken after 16 h clearly showed that nuclei in hypocotyl cells of the dark-grown seedling had a relatively larger shift in green fluorescence as compared to hypocotyl cells maintained in light (Fig. 4, C and D). The increased green fluorescence of nuclei in dark-grown seedlings (Fig. 4D) suggested a net increase in their H2B content and this correlated well with the change in cell length (Fig. 4F). Interestingly, although the cell size did not change much in light-grown seedlings (Fig. 4E) their nuclei also showed a slight shift in their green fluorescence. This shift suggests that there is an increase of fresh H2B::mEosFP; however, it should be noted that the color shift observed in dark-grown seedlings is unquestionably greater than in light-grown seedlings (Fig. 4, D versus C). Clearly other factors such as the use of a constitutively active CaMV-35S promoter in this study or the gradual degradation of the photoconverted form of mEosFP might have also played a role in creating this slight shift. Recognizing this discrepancy our subsequent efforts are directed to using the native H2B promoter and comparing the resultant transgenic plants with our prom35S transgenics. Nonetheless these observations reaffirmed a link between increased nuclear activity and cell expansion.
Figure 4.
Cell size and color change in nuclei of light- and dark-grown hypocotyl cells of transgenic Arabidopsis plants expressing H2B::mEosFP. A and B (section 1), Green fluorescent state of nuclei visible in seedlings grown in light. Section 2 shows the same seedlings irradiated with violet-blue light for 6 × 45 s to photoconvert nuclei into exhibiting red fluorescence. Subsequently the seedling in A was maintained in light while seedling B was kept in dark. Section 3 shows the two seedlings 16 h after photoconversion. The red color of nuclei is maintained in the light-grown seedling but nuclei in the dark-grown seedling shift to green. C and D, A scatter plot depicting green and red fluorescence values of individual nuclei in A (1–3) and B (1–3) confirms the shift toward green color that occurs in nuclei of dark-grown seedling (B, 3) in comparison to the nuclei of light-grown seedling (A, 3). E and F, Measurement of hypocotyl cells of light- (E) and dark-treated (F) seedlings shows that during the 16 h time that color change occurs in nuclei, the cells in dark-grown hypocotyl also undergo considerable elongation. Size bar in A and B = 100 μm.
Whereas technical limitations in long-term imaging of developing plants have not allowed us to observe multiple endocycles in the same cell so far, the proof of concept experiments presented here unequivocally demonstrate that photoconvertible H2B::mEosFP can be used to detect changes in nuclear activity underlying developmental events. In principle, once long-term imaging of individual nuclei has become possible, reiterations of photoconversion and color recovery can be used to chart the time of start and progression through multiple endocycling events. Unlike any other nuclear probes available so far the use of H2B::mEosFP entails no tissue maceration or fixation and allows direct observations in living plant cells. Photoconvertible H2B::mEosFP and the stable transgenic lines expressing it are valuable additions to the tool box available for dissecting nuclear behavior and the occurrence of polysomaty in plants.
MATERIALS AND METHODS
Molecular Methods
The histone H2B cDNA (At3g45980) was PCR amplified using primers forward JM-258 (TCTAGAGGTATGGCGCCGAGAGCAGAGAAG) and reverse JM-259 (GGATCCGATTCCAGAGCTTGTGAATTTGGTA) that introduced XhoI and BamHI restriction sites at the 5′/3′, respectively, while removing the stop codon. The 453-bp fragment was ligated with mEosFP (Mathur et al., 2010) at the BamHI site to finally produce p35SCaMV-H2B::mEosFP-pA in the pCAMBIA 1300 binary vector (http://www.cambia.org.au) to which a pCaMV-35S promoter and a nos terminator sequence had been added. Standard molecular biology protocols were followed for the cloning (Sambrook et al., 1989).
Expression in Plant Cells
Transient expression of pro35SCaMV-H2B::mEosFP-pA was carried out through agroinfiltration (Agrobacterium tumefaciens strain GV3101 at an optical density of 0.8 at 600 nm) of tobacco (Nicotiana tabacum) leaves according to Sparkes et al. (2006). Images of H2B::mEosFP expression were taken between 24 and 48 h after infiltration. Stable transgenic lines for H2B::mEosFP were generated using A. tumefaciens-mediated floral-dip transformation method (Clough and Bent, 1998). Transgenic hygromycin-resistant seeds were grown on 1% agar-gelled Murashige and Skoog (1962) medium supplemented with 3% Suc and with pH adjusted to 5.8. Plants were grown in petri dishes in an incubator maintained at 21°C ± 2°C and a 16-/8-h light/dark regime using cool-white light of approximately 80 to 100 μmol m−2 s−2.
Tobacco BY2 cells (Nagata et al., 2003) expressing H2B::mEosFP were obtained through cocultivation with A. tumefaciens carrying the probe followed by hygromycin selection. Stable transgenic lines were maintained through weekly subculture using standard protocol for BY2 cells (Kumagai-Sano et al., 2007).
Microscopy
Imaging Chambers
To capture the early stages of collet and root hair formation over several hours, shallow germinating chambers were created. These contained Murashige and Skoog medium gelled with 1% agarose poured onto the lid (90 mm) of a plastic petri dish. A seed was placed at the rim of the medium cushion. To maintain free space for the developing seedling two glass coverslips (approximately 0.17-mm thick) were placed as spacers on both sides of the seed and yet another coverslip (24 × 60 mm) was used as cover. The petri dish containing chambers were sealed with parafilm and placed at 4°C to break seed dormancy. After 48 h cold treatment chambers were incubated upright for additional 24 to 48 h in a standard growth chamber. Seedlings growing in the chamber were usually imaged soon after the emergence of the radicle. Long-term imaging of BY2 cells was carried out by observing cells placed in the middle of a circular chamber created by painting silicon grease and covering with a glass coverslip. A small gap was left in the seal to allow gaseous exchange.
Photoconversion and Imaging
Photoconversion was varied according to the brightness of the nucleus and was carried out by illuminating young seedlings with violet-blue light in three to seven cycles of 45-s illumination with 15-s breaks, unless otherwise stated, using a standard HBO 100 W/2 mercury short arc lamp and the Leica fluorescence filter set D (excitation filter 355–425 nm; dichromatic mirror 455 nm; suppression filter LP 470 nm). The epifluorescence setup consisted of a Leica DM-6000CS microscope. Photoconversion was performed manually by controlling the diaphragm as described earlier (Mathur et al., 2010). Subsequent simultaneous imaging of unphotoconverted and photoconverted H2B::mEosFP was carried out using a Leica TCS-SP5 confocal laser-scanning unit equipped with a 488-nm argon and a 543-nm helium-neon laser. To avoid photobleaching of unphotoconverted green mEosFP the 488-nm laser intensity was kept at a minimum. For visualizing H2B::mEosFP, the probe was excited using a 488- and a 458-nm laser, and emissions collected between 511 and 540 nm for unphotoconverted green mEosFP, and between 568 and 600 nm for photoconverted red mEosFP. All images were captured using at a color depth of 24-bit RGB. The excitation, emission, black level, and pinhole settings were rigorously maintained during confocal imaging throughout a time-lapse experiment. Fluorescent microspheres (Focal Check; 6-μm diameter; Molecular Probes-F14807) were used as standard control for ensuring conformity between experiments on red fluorescence stability in photoconverted guard cell nuclei.
Postacquisition Image Processing and Quantification
All color measurements were carried out using 24-bit RGB images without any prior image manipulations except for maximum intensity projections of confocal z stacks. Due to the green to red photoconversion and recovery of green by mEosFP expression every nucleus is characterized by a combination of a RG value. The RG color characteristics of single nuclei were often depicted through scatter plots (e.g. Figs. 2G and 4, C and D). The average RG pixel brightness values of single nuclei for these plots were measured with the image-processing software FIJI (http://fiji.sc/wiki/index.php/Fiji) by encircling the respective nucleus with the selection tools and applying the color histogram command that provides characteristic RG values, each distributed onto a 0 to 255 scale. In the plots a change in fluorescence value results in a change in the position of respective data point in the x/y dimensions of the scatter plot.
To follow changes in color over time (e.g. Fig. 3E), nuclei were tracked over time and their RG values measured as described above. To reduce data dimensionality and create a two-dimensional plot, green to red ratios were calculated and plotted against time.
For testing a potential connection between color, nucleus size, and distribution in the seedling a single frame (e.g. 1,760-min time point; Fig. 3E) of a time series of a developing seedling (e.g. Supplemental Movie S1) was chosen. For an estimate of the nuclear size in addition to RG values, the area of the nuclei, which is visible in the maximum intensity z-projected images, was measured using the measure command of Fiji. To build a histogram of the color distribution among the nuclei represented in the field of view the basic statistics command of StatPlus (www.analystsoft.com) was used. Box plots, scatter plots, as well as Student’s t test were performed using SigmaPlot (Systat Software, Inc.) or R (http://www.r-project.org/). Regression analysis was carried out using the Statistical Analysis Software (SAS; http://www.sas.com/technologies/analytics/statistics/stat/index.html>).
For presentation and assembly of microscopic images all images were cropped and processed for brightness/contrast as complete montages using Adobe Photoshop CS3 and Adobe Illustrator CS4 (http://www.adobe.com). The layer function of this software was used to introduce text, regions of interest, and color overlays.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Measurements of red fluorescence intensity in guard cells.
Supplemental Figure S2. Location of subpopulation of nuclei in developing hypocotyl and collet hair cells.
Supplemental Figure S3. Internal controls for ensuring stability of image acquisition parameters during visualization of nuclei.
Supplemental Movie S1. Time-lapse image series of a germinating, photoconverted H2B::mEosFP seedling starting at 720 min after photoconversion and ending at 1,760 min (60-min step size).
Acknowledgments
We thank Joerg Wiedenmann for mEosFP and Rob Mullen for tobacco BY2 cells.
References
- Adachi S, Minamisawa K, Okushima Y, Inagaki S, Yoshiyama K, Kondou Y, Kaminuma E, Kawashima M, Toyoda T, Matsui M, et al. (2011) Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc Natl Acad Sci USA 108: 10004–10009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barow M. (2006) Endopolyploidy in seed plants. Bioessays 28: 271–281 [DOI] [PubMed] [Google Scholar]
- Berger CA. (1941) Reinvestigation of polysomaty in spinacia. Bot Gaz 102: 759–769 [Google Scholar]
- Boisnard-Lorig C, Colon-Carmona A, Bauch M, Hodge S, Doerner P, Bancharel E, Dumas C, Haseloff J, Berger F. (2001) Dynamic analyses of the expression of the HISTONE:YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13: 495–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudonck K, Dolan L, Shaw PJ. (1999) The movement of coiled bodies visualized in living plant cells by the green fluorescent protein. Mol Biol Cell 10: 2297–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramsiepe J, Wester K, Weinl C, Roodbarkelari F, Kasili R, Larkin JC, Hülskamp M, Schnittger A. (2010) Endoreplication controls cell fate maintenance. PLoS Genet 6: e1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer C, Ishida T, Sugimoto K. (2010) Developmental control of endocycles and cell growth in plants. Curr Opin Plant Biol 13: 654–660 [DOI] [PubMed] [Google Scholar]
- Castellano MdelM, Boniotti MB, Caro E, Schnittger A, Gutierrez C. (2004) DNA replication licensing affects cell proliferation or endoreplication in a cell type-specific manner. Plant Cell 16: 2380–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro S, Loureiro J, Rodriguez E, Silveira P, Navarro L, Santos C. (2007) Evaluation of polysomaty and estimation of genome size in Polygala vayredae and P. calcarea using flow cytometry. Plant Sci 172: 1131–1137 [Google Scholar]
- Chiu WL, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Chytilova E, Macas J, Galbraith DW. (1999) Green fluorescent protein targeted to the nucleus, a transgenic phenotype useful for studies in plant biology. Ann Bot (Lond) 83: 645–654 [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin SG, Marco Y. (2003) Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA 100: 8024–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Fang Y, Spector DL. (2005) Centromere positioning and dynamics in living Arabidopsis plants. Mol Biol Cell 16: 5710–5718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fass E, Shahar S, Zhao J, Zemach A, Avivi Y, Grafi G. (2002) Phosphorylation of histone h3 at serine 10 cannot account directly for the detachment of human heterochromatin protein 1gamma from mitotic chromosomes in plant cells. J Biol Chem 277: 30921–30927 [DOI] [PubMed] [Google Scholar]
- Fong PM, Tian L, Chen ZJ. (2006) Arabidopsis thaliana histone deacetylase 1 (AtHD1) is localized in euchromatic regions and demonstrates histone deacetylase activity in vitro. Cell Res 16: 479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foudi A, Hochedlinger K, Van Buren D, Schindler JW, Jaenisch R, Carey V, Hock H. (2009) Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat Biotechnol 27: 84–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Knapp S. (1991) Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol 96: 985–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gindullis F, Peffer NJ, Meier I. (1999) MAF1, a novel plant protein interacting with matrix attachment region binding protein MFP1, is located at the nuclear envelope. Plant Cell 11: 1755–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann K, Evans DE. (2011) Nuclear envelope dynamics during plant cell division suggest common mechanisms between kingdoms. Biochem J 435: 661–667 [DOI] [PubMed] [Google Scholar]
- Graumann K, Runions J, Evans DE. (2010) Characterization of SUN-domain proteins at the higher plant nuclear envelope. Plant J 61: 134–144 [DOI] [PubMed] [Google Scholar]
- Grebenok RJ, Pierson E, Lambert GM, Gong FC, Afonso CL, Haldeman-Cahill R, Carrington JC, Galbraith DW. (1997) Green-fluorescent protein fusions for efficient characterization of nuclear targeting. Plant J 11: 573–586 [DOI] [PubMed] [Google Scholar]
- Hardin JA, Einem GE, Lindsay DT. (1967) Simultaneous synthesis of histone and DNA synchronously dividing Tetrahymena pyriformis. J Cell Biol 23: 709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnilica LS. (1972) The Structure and Biological Function of Histones. CRC Press, Cleveland, OH, pp 1–213 [Google Scholar]
- Hülskamp M, Misŕa S, Jürgens G. (1994) Genetic dissection of trichome cell development in Arabidopsis. Cell 76: 555–566 [DOI] [PubMed] [Google Scholar]
- Ingouff M, Hamamura Y, Gourgues M, Higashiyama T, Berger F. (2007) Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr Biol 17: 1032–1037 [DOI] [PubMed] [Google Scholar]
- Ingouff M, Rademacher S, Holec S, Soljić L, Xin N, Readshaw A, Foo SH, Lahouze B, Sprunck S, Berger F. (2010) Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr Biol 20: 2137–2143 [DOI] [PubMed] [Google Scholar]
- Inzé D, De Veylder L. (2006) Cell cycle regulation in plant development. Annu Rev Genet 40: 77–105 [DOI] [PubMed] [Google Scholar]
- Irons SL, Evans DE, Brandizzi F. (2003) The first 238 amino acids of the human lamin B receptor are targeted to the nuclear envelope in plants. J Exp Bot 54: 943–950 [DOI] [PubMed] [Google Scholar]
- Ishida T, Adachi S, Yoshimura M, Shimizu K, Umeda M, Sugimoto K. (2010) Auxin modulates the transition from the mitotic cycle to the endocycle in Arabidopsis. Development 137: 63–71 [DOI] [PubMed] [Google Scholar]
- Ishida T, Kurata T, Okada K, Wada T. (2008) A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol 59: 365–386 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Schnittger A. (2004) Cell cycle and differentiation. Curr Opin Plant Biol 7: 661–669 [DOI] [PubMed] [Google Scholar]
- John PCL, Qi R. (2008) Cell division and endoreduplication: doubtful engines of vegetative growth. Trends Plant Sci 13: 121–127 [DOI] [PubMed] [Google Scholar]
- Jovtchev G, Schubert V, Meister A, Barow M, Schubert I. (2006) Nuclear DNA content and nuclear and cell volume are positively correlated in angiosperms. Cytogenet Genome Res 114: 77–82 [DOI] [PubMed] [Google Scholar]
- Kim SH, Ryabov EV, Kalinina NO, Rakitina DV, Gillespie T, MacFarlane S, Haupt S, Brown JWS, Taliansky M. (2007) Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J 26: 2169–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H. (2005) Histone dynamics in living cells revealed by photobleaching. DNA Repair (Amst) 4: 939–950 [DOI] [PubMed] [Google Scholar]
- Kimura H, Cook PR. (2001) Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J Cell Biol 153: 1341–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondorosi E, Roudier F, Gendreau E. (2000) Plant cell-size control: growing by ploidy? Curr Opin Plant Biol 3: 488–492 [DOI] [PubMed] [Google Scholar]
- Kumagai-Sano F, Hayashi T, Sano T, Hasezawa S. (2007) Cell cycle syncronization of tobacco BY-2 cells. Nat Protoc 1: 2621–2627 [DOI] [PubMed] [Google Scholar]
- Lee HO, Davidson JM, Duronio RJ. (2009) Endoreplication: polyploidy with purpose. Genes Dev 23: 2461–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Tessadori F, Germann S, Snijder B, Fransz P, Gaudin V. (2005) The Arabidopsis LHP1 protein is a component of euchromatin. Planta 222: 910–925 [DOI] [PubMed] [Google Scholar]
- Lindhout BI, Fransz P, Tessadori F, Meckel T, Hooykaas PJJ, van der Zaal BJ. (2007) Live cell imaging of repetitive DNA sequences via GFP-tagged polydactyl zinc finger proteins. Nucleic Acids Res 35: e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković ZJ, Hilscher J, Barta A. (2004) Use of fluorescent protein tags to study nuclear organization of the spliceosomal machinery in transiently transformed living plant cells. Mol Biol Cell 15: 3233–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Radhamony R, Sinclair AM, Donoso A, Dunn N, Roach E, Radford D, Mohaghegh PSM, Logan DC, Kokolic K, et al. (2010) mEosFP-based green-to-red photoconvertible subcellular probes for plants. Plant Physiol 154: 1573–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melaragno JE, Mehrotra B, Coleman AW. (1993) Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5: 1661–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes ICR, Lermontova I, Schubert I. (2011) Recognition of A. thaliana centromeres by heterologous CENH3 requires high similarity to the endogenous protein. Plant Mol Biol 75: 253–261 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nagata T, Sakamoto K, Shimizu T. (2003) Tobacco BY-2 cells: the present and beyond. In Vitro Cell Dev Biol Plant 40: 163–166 [Google Scholar]
- Pay A, Resch K, Frohnmeyer H, Fejes E, Nagy F, Nick P. (2002) Plant RanGAPs are localized at the nuclear envelope in interphase and associated with microtubules in mitotic cells. Plant J 30: 699–709 [DOI] [PubMed] [Google Scholar]
- Pih KT, Yi MJ, Liang YS, Shin BJ, Cho MJ, Hwang I, Son D. (2000) Molecular cloning and targeting of a fibrillarin homolog from Arabidopsis. Plant Physiol 123: 51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins E, Borun TW. (1966) The cytoplasmic synthesis of histones in HeLa cells and its temporal relationship to DNA replication. Proc Natl Acad Sci USA 57: 409–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder AHK, Chickarmane V, Cunha A, Obara B, Manjunath BS, Meyerowitz EM. (2010) Variability in the control of cell division underlies sepal epidermal patterning in Arabidopsis thaliana. PLoS Biol 8: e1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A, Meier I. (2001) A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc Natl Acad Sci USA 98: 15377–15382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Ed 2 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sawchuk MG, Head P, Donner TJ, Scarpella E. (2007) Time-lapse imaging of Arabidopsis leaf development shows dynamic patterns of procambium formation. New Phytol 176: 560–571 [DOI] [PubMed] [Google Scholar]
- Shaner NC, Patterson GH, Davidson MW. (2007) Advances in fluorescent protein technology. J Cell Sci 120: 4247–4260 [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C. (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Roberts GR, Stacey NJ, McCann MC, Maxwell A, Roberts K. (2005) RHL1 is an essential component of the plant DNA topoisomerase VI complex and is required for ploidy-dependent cell growth. Proc Natl Acad Sci USA 102: 18736–18741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Roberts K. (2003) “Big it up”: endoreduplication and cell-size control in plants. Curr Opin Plant Biol 6: 544–553 [DOI] [PubMed] [Google Scholar]
- Tamura K, Fukao Y, Iwamoto M, Haraguchi T, Hara-Nishimura I. (2010) Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell 22: 4084–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Hara-Nishimura I. (2011) Involvement of the nuclear pore complex in morphology of the plant nucleus. Nucleus 2: 168–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bruaene N, Joss G, Thas O, Van Oostveldt P. (2003) Four-dimensional imaging and computer-assisted track analysis of nuclear migration in root hairs of Arabidopsis thaliana. J Microsc 211: 167–178 [DOI] [PubMed] [Google Scholar]
- Van Damme D, Bouget FY, Van Poucke K, Inzé D, Geelen D. (2004) Molecular dissection of plant cytokinesis and phragmoplast structure: a survey of GFP-tagged proteins. Plant J 40: 386–398 [DOI] [PubMed] [Google Scholar]
- Walker JD, Oppenheimer DG, Concienne J, Larkin JC. (2000) SIAMESE, a gene controlling the endoreduplication cell cycle in Arabidopsis thaliana trichomes. Development 127: 3931–3940 [DOI] [PubMed] [Google Scholar]
- Wang H, Ma LG, Li JM, Zhao HY, Deng XW. (2001) Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154–158 [DOI] [PubMed] [Google Scholar]
- Wiedenmann J, Ivanchenko S, Oswald F, Schmitt F, Röcker C, Salih A, Spindler K-D, Nienhaus GU. (2004) EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc Natl Acad Sci USA 101: 15905–15910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HQ, Tang RH, Cashmore AR. (2001) The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13: 2573–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Sayegh R, Maymon M, Warpeha K, Klejnot J, Yang H, Huang J, Lee J, Kaufman L, Lin C. (2009) Formation of nuclear bodies of Arabidopsis CRY2 in response to blue light is associated with its blue light-dependent degradation. Plant Cell 21: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Li Y, Ben-Meir H, Oliva M, Mosquna A, Kiss V, Avivi Y, Ohad N, Grafi G. (2006) Different domains control the localization and mobility of LIKE HETEROCHROMATIN PROTEIN1 in Arabidopsis nuclei. Plant Cell 18: 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Gong FC, Lambert GM, Galbraith DW. (2005) Cell type-specific characterization of nuclear DNA contents within complex tissues and organs. Plant Methods 1: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li X. (2005) A putative nucleoporin 96 is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1,constitutive 1. Plant Cell 17: 1306–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]