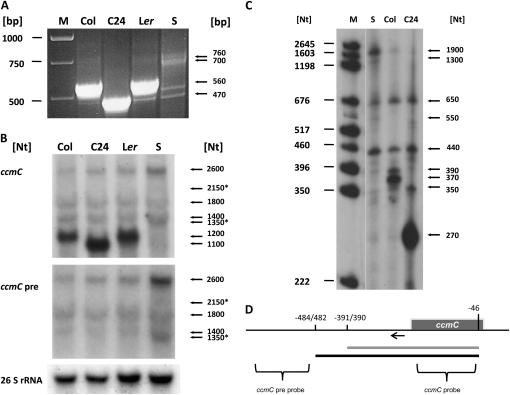
Figure 1.
Analysis of ccmC transcripts. RNAs derived from the ccmC gene in accession Col, C24, and Ler and the mutant line SAIL 18E04 (S) were analyzed by CR-RT-PCR (A), northern-blot hybridization (B), and primer extension analysis (C). Lengths of size markers are given on the left-hand sides of the images. Relevant products or signals are indicated by small arrows and corresponding sizes on the right-hand sides. Asterisks indicate mRNA species predominantly detected in the mutant (the 2.15-kb mRNA is difficult to see in the image, but can be seen on the x-ray film). D, Schema of the ccmC gene (dark-gray box) and the major transcripts (gray [C24] and black [Col] bold lines). Mature 5′ (relative to the translation start codon NATG, n = −1) and 3′ ends (relative to the translation stop codon, TAAN, n = +1) are indicated. Probes used for the northern analysis are depicted in the bottom part of the schema and indicated on the left-hand side of the images in B. The position of the oligonucleotide (Atccb3Mega5′.nah) used for the primer extension experiments is indicated by an arrow. Schema not drawn to scale.