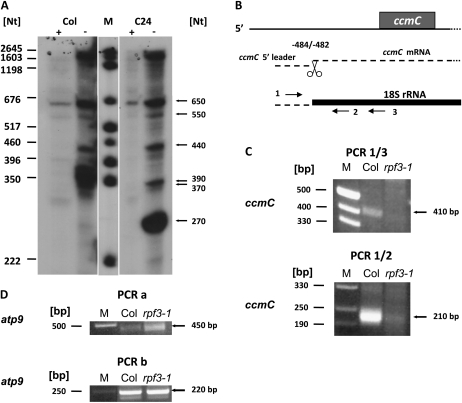
Figure 4.
The mature ccmC 5′ ends are generated by posttranscriptional processing. A, Primer extension analysis of untreated (−) and terminator exonuclease-treated (+) mtRNA from Col and C24. B, Schema explaining the analysis of 3′ ends of potential ccmC 5′ leader molecules. The ccmC reading frame is indicated by a gray box, ccmC RNAs are given as dashed lines. The 18S rRNA, which was used as anchor molecule, is depicted as a black bold line. Primers (horizontal arrows) used for this analysis are Atccb3QPCR.3 (1), At18S-Adapter.R4 (2), and At18S-Adapter.R3 (3). C, PCR products amplified with primer pair 1/3 (top section) and 1/2 (bottom section) from Col wild type and from rpf3-1 total mtRNA. Lengths of size markers are given on the left, relevant products are indicated on the right-hand side of the images (A and C). D, Control reactions performed with primer pairs Atatp9Mega-Endo.A/At18S-Adapter.R3 (PCR a) and Atatp9-11/At18S-Adapter.R4 (PCR b) amplified atp9 leader RNA molecules as described previously (Forner et al., 2007). These reactions were performed with identical volumes of the same cDNA preparation used in the PCRs shown in C. The results of these amplifications demonstrate that in rpf3-1 the reduced amount of PCR product derived of ccmC leader molecules is not due to the use of lower amounts or insufficient quality of the cDNA.