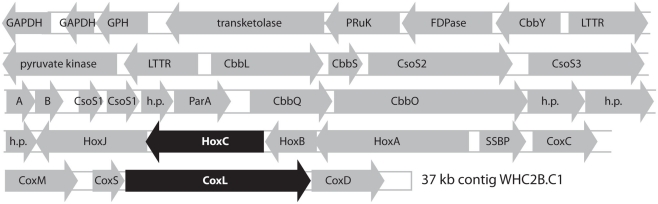
Figure 4.
Predicted products of open reading frames identified in the longest contig from Tablelands fluid sample WHC2b (37 kb, 67% GC, comprising 615 shotgun reads at 6.6 mean coverage). The putative large subunits of [NiFe]-hydrogenase (HoxC) and carbon monoxide dehydrogenase (CoxL) are highlighted in black to indicate that their phylogenetic relationships are shown in Figures 5 and Figure A1 in Appendix. GAPDH: NAD-dependent glyceraldehyde-3-phosphate dehydrogenase. GPH, phosphoglycolate phosphatase; PRuK, phosphoribulokinase; FDPase, type I fructose-1,6-bisphosphatase; CbbY, rubisco-associated hydrolase; LTTR, LysR-type transcriptional regulator; CbbL, CbbS, ribulose bisphosphate carboxylase, large and small chains; CbbS, ribulose bisphosphate carboxylase small chain; CsoS2, CsoS3, CsoS1, carboxysome shell proteins; A, B, putative carboxysome peptides; h. p., hypothetical protein; ParA, chromosome (plasmid) partitioning protein; CbbQ, CbbO, rubisco activation proteins; HoxJ, HoxA, hydrogenase regulation proteins; HoxC, HoxB, uptake [NiFe]-hydrogenase, large and small subunits; SSBP, single-stranded DNA binding protein; CoxC, carbon monoxide dehydrogenase signaling protein; CoxM, CoxS, CoxL, carbon monoxide dehydrogenase, medium, small, and large subunits; CoxD, carbon monoxide dehydrogenase accessory protein.