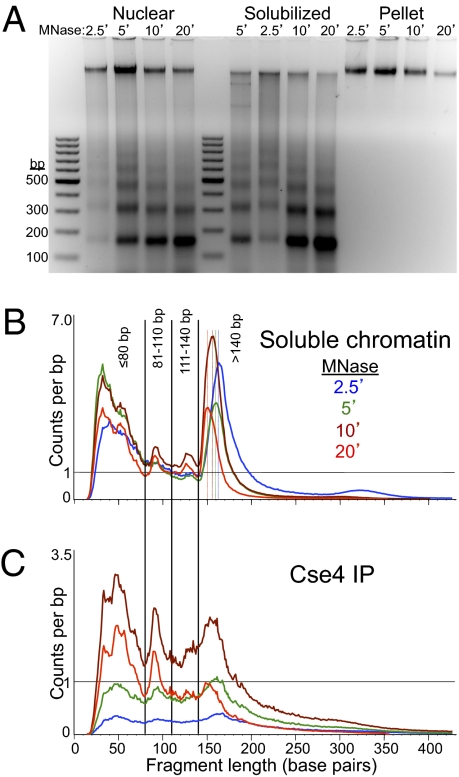
Fig. 1.
Paired-end sequencing of soluble chromatin and Cse4 ChIP yields distinct size classes of MNase-protected particles. (A) Agarose gel analysis of MNase time-point samples showing DNA from whole nuclei extracted from strain SBY5146 after MNase digestion (Nuclear), DNA from soluble chromatin after needle extraction and pooling of extracts (Solubilized), and DNA from the insoluble residue after solubilization (Pellet). The gradual reduction in size of protected DNA fragments can be seen as jogs in the solubilized chromatin samples loaded out-of-order (5, 2.5, 10, and 20 min) in the middle set of lanes. Size distributions of mapped paired-end reads for Solubilized chromatin (B) and FLAG-Cse4 ChIP (C), showing the size classes chosen for further analysis.