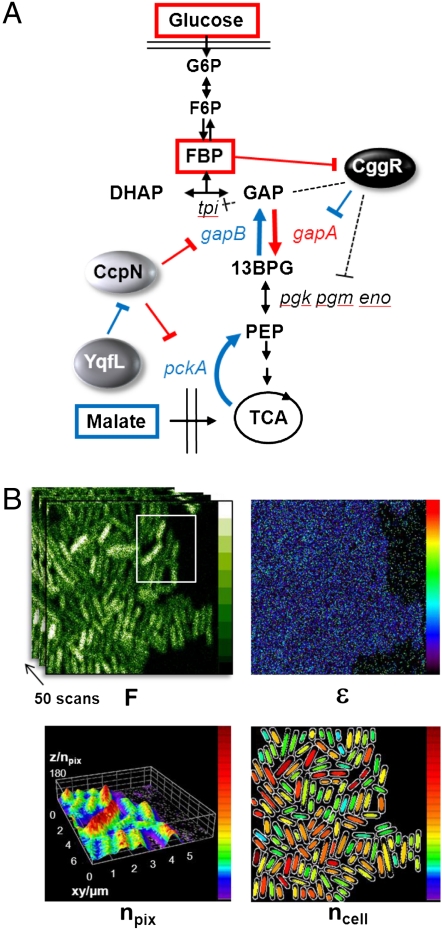
Fig. 1.
(A) Schematic of the central carbon metabolism showing the switch between glycolysis and gluconeogenesis controlled by the repressors CggR and CcpN. Important metabolites are in squares, regulatory proteins are ellipses, and the genes coding for the enzymes are in small italic letters. When glucose is available for cell growth, fructose-1,6-biphosphate (FBP) accumulates and blocks the repressive action that CggR exerts on the transcription of gapA and four other central glycolytic genes (pgk, pgm, eno, and tpi). Because CggR is transcribed from the same gapA operon that it represses, it is also an autorepressor. Inversely, when cells are grown on malate or other nonglycolytic carbon sources, the CcpN repressor is inhibited by an unknown mechanism involving YqfL, allowing expression of the essential gluconeogenic genes gapB and pckA. (B) Schematic of 2psN&B experiments. A stack of 50 raster scans of agarose immobilized live cells of B. subtilis expressing gfpmut3 are recorded using infrared (930 nm) laser excitation and a dwell time of 50 μs at each pixel (faster than GFP diffusion); full scale of fluorescence intensity (F) is 10 photon counts/pixel/50 μs laser dwell time. The fluorescence fluctuations relative to the mean at each pixel are used to calculate the pixel-based maps of the true (shot noise corrected) molecular brightness (ϵ, full scale 1 photon/molecule/50 μs dwell time) and the number (npix) of the fluorescent particles detected in the 2-photon excitation volume (volex = 0.07 fL inside B. subtilis); a 3D surface plot of npix is shown for the white-delineated area of the above intensity panel. Bottom right: Cartoon representation of the individual cells auto-detected using PaTrack (40) and showing the 50% central pixels used for averaging the particles number in each cell (ncell); the full scale for the npix and ncell maps is 180 molecules/volex.