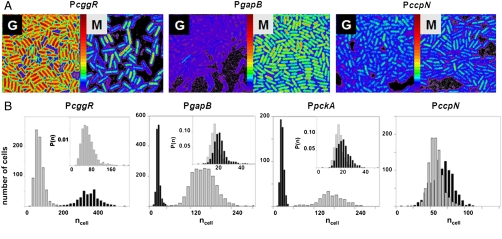
Fig. 2.
Cell-by-cell quantification of catabolite regulation in B. subtilis by 2psN&B. (A) Pixel-based fluorescent particles number maps of B. subtilis cells expressing gfpmut3 transcriptional fusion from PccgR, PgapB (results are similar for PpckA; not shown), and PccpN. Cells harvested from liquid cultures containing 0.5% glucose (G) or 0.5% malate (M) as the sole carbon source were immobilized on agarose pads for 2psN&B analysis as described in Fig. 1B. The full scale is 360 molecules/volex. (B) Cell-based particles number (ncell) distributions for the indicated promoter-gfpmut3 fusion strains grown on glucose (black) or malate (gray). Inset in the first panel shows the expanded histogram of the probability density function P(ncell) measured in malate for PcggR. Insets in panel 2 and 3 show the expanded histogram of the probability density function P(ncell) observed in glucose for the PgapB and PpckA promoters in black, and that observed for the background BSB168 strain in gray.